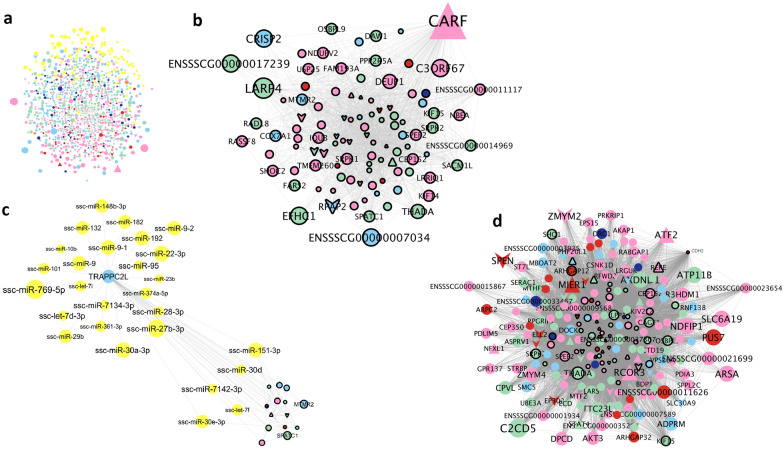
Fig. 3.
Co-association network based on the AWM and transcriptomics data. a Full network with 1313 genes and 94 miRNAs; b Subset of the network showing the transcription factor CARF and all its predicted interactions; c Subset of the network with the TRAPPC2L interactions, which included several miRNAs; d Subset of the network with the CHD2 gene interactions. The node color corresponds to the phenotype group with the highest correlation value, as follows: concentration (red), abnormal acrosomes (green), abnormalities and droplets (pink), osmotic resistance test (orange), motility (light blue) and viability (dark blue). miRNAs are depicted in yellow. Node and text sizes correspond to the number of significant phenotypes correlated with that gene or miRNA. Nodes with a black line border correspond to genes identified in the shared network. Node shape indicates classification as: triangle (TF), V (TF co-factor) and ellipse (other genes and miRNAs)