Fig. 4.
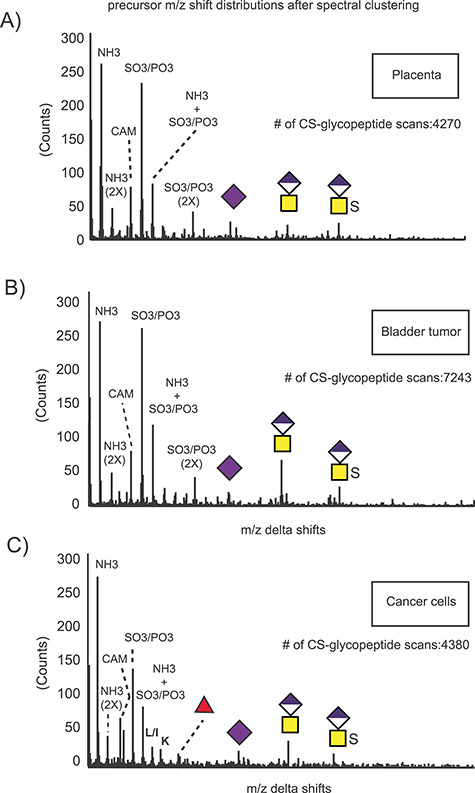
Glycoform analysis of linkage region substitutions through spectral clustering and molecular networking. All MS/MS scans were filtered based on the presence of the CS-specific oxonium ion m/z 362.11, and the selected glycopeptide scans were clustered based on spectral similarities. The m/z precursor shift distributions for the clustered glycopeptides derived from the human placenta (A), bladder tumor (B) and cancer cell lines (C) reveal the presence of structural variations in the samples based on number of sulfate groups, length of the depolymerized linkage regions, sialic acid and fucose as well as artefactual chemical modifications such as ammonium adducts and carbamidomethylation.
