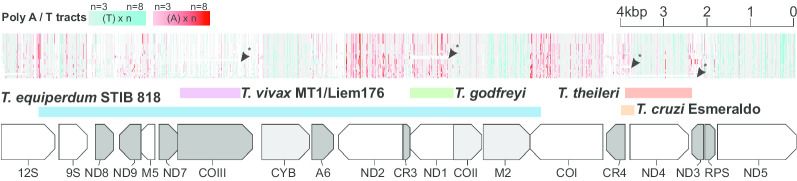
Fig. 1.
Global overview of maxicircle gene coding region. Top: alignment of the maxicircle mitochondrial DNA gene coding regions from 27 isolates (top to bottom, Trypanosoma brucei H866, 1829 ALJO, Lister 427, TREU 927, TSW 55, J10, LF1; T. congolense IL3000, WG81, GAM2, IL3900, IL3578, ERA D1; T. simiae ERA C2; T. godfreyi KEN7, ERA F1; T. vivax Liem 176, Y486, Tv2323, T. cruzi, CL Brener, Esmeraldo; T. lewisi, T. conorhini, T. copemani, T. grayi, T. theileri, Leishmania tarentolae). Tracts of poly-T or poly-A are shown coloured turquoise or red respectively. An approximate scale is shown. Segmental gene deletions in the alignment are indicated by arrows and are also shown below as coloured bars; the deletion from T. equiperdum STIB 818 is also shown for comparison. Bottom: gene order in the maxicircle gene coding region. Non-edited genes in are shown in white, minimally edited genes in light grey and extensively edited (pan-edited) genes in grey. Editing categories are on the basis of T. vivax [14]