Figure 2.
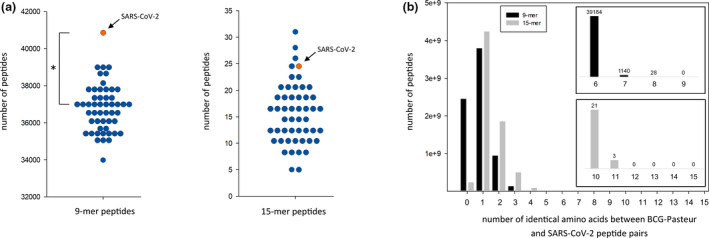
Exploration of peptide similarity between the Bacillus Calmette–Guérin (BCG)‐Pasteur proteome and the severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) proteome. (a) Comparison of the BCG‐Pasteur proteome to the SARS‐CoV‐2 proteome and randomised proteomes. 9‐mer and 15‐mer peptides of the BCG‐Pasteur proteome were mapped to the SARS‐CoV‐2 proteome and 50 random proteomes containing the same number and length proteins as the original SARS‐CoV‐2 proteome but with scrambled amino acid composition. The number of 9‐mer and 15‐mer BCG‐Pasteur peptides ≥ 67% identical to a SARS‐CoV‐2 peptide are shown. Interquartile range analysis of outliers identified values higher than 40 222 as an outlier for 9‐mer sequences and 30.25 for 15‐mer sequences. According to this analysis, the SARS‐CoV‐2 9‐mer value (40 848) was an outlier, but the SARS‐CoV‐2 15‐mer value (24) was not. Grubbs' test for outliers was applied to calculate the significance of the 40 848 value as an outlier. *P < 0.05. (b) Exploration of the identity between BCG‐Pasteur peptides and SARS‐CoV‐2 peptides. 9‐mer and 15‐mer BCG‐Pasteur peptides were mapped to the SARS‐CoV‐2 proteome. The number of BCG‐Pasteur peptides with different level of identity to a SARS‐CoV‐2 peptide is shown. Inset graphs show the occurrence of the 9‐mer and 15‐mer peptides with ≥ 67% identity.
