Figure 6. Comparison of phage‐derived ligands and natural SLiMs in complex with PRMs.
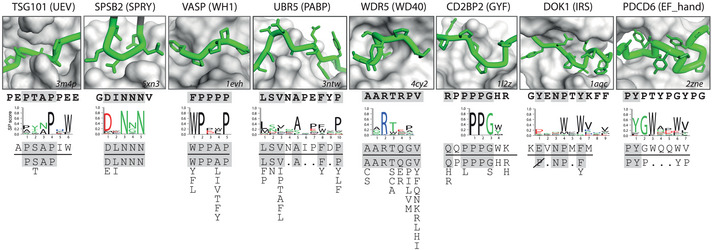
Depicted are eight PRMs for which phage‐derived specificity profiles were determined (Fig 1) and for which natural SLiM ligand information is available in the ELM repository (Dataset EV6). The name of the protein from which the studied PRM was derived is listed at the top with the PRM family name in parenthesis. The box shows a close‐up of the structure of the peptide‐binding site of the PRM, which is depicted as a gray surface (the PDB entry code is shown in the bottom right corner). The peptide ligand is colored green with the main chain shown as a tube and side chains shown as sticks (the N terminus is to the left), and its sequence is shown directly below the structure in bold text. Below the peptide ligand sequence, the following are shown: the phage‐derived specificity profile, the phage‐derived peptide most similar to the ELM motif, and the ELM motif (below the horizontal line). Sequences in the peptide structure, the peptide ligand, and the phage‐derived peptide that match the ELM motif are shaded gray. The ELM motifs are arranged vertically to align with the specificity profile and peptides. Position allowing any amino acid except Pro is depicted as “P” crossed out with a diagonal line.
