Figure 3.
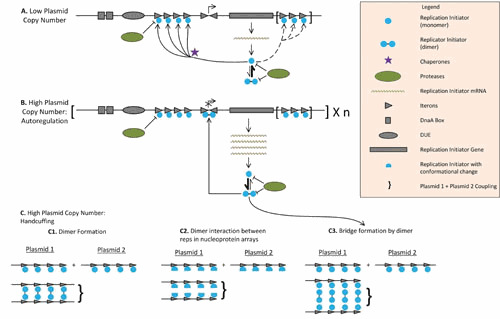
Replication initiator proteins as negative regulators of replication. The rate of replication is determined by the concentration of free (non-Rep-bound) iterons, which is determined by both plasmid copy number and cell volume. Given the cooperative interactions involved in saturating iterons, this regulatory mechanism is ultrasensitive to plasmid copy number and switch-like. Generic class A theta replicon structure: rep gene (rectangle), DnaA boxes, which can be found upstream or downstream of the Rep (squares), iterons (arrowheads), and an AT-rich DNA unwinding element (DUE, oval). (A) Replication initiator proteins (Reps) when the plasmid copy number is low. Reps (blue circles) are expressed from strong promoters and tend to be in their monomeric (active) form; in some cases, the activation of these monomers needs to be facilitated by chaperones (purple stars). Reps are also sensitive to protease activity (green ovals). Active monomers bind iterons until saturation; in some cases, additional clusters of iterons are present to decrease the level of Rep available, further tightening plasmid copy number control (86). (B) Replication initiator proteins when plasmid copy number is high. High Rep protein expression resulting from a high plasmid copy number favors the dimeric form of Rep. The symmetrical conformation of the dimeric form matches inverted iterons found in the promoter of rep genes. This results in a binding affinity for the promoter that is higher than that of the RNA polymerase, blocking rep transcription (137). Transcriptional autorepression by Reps is seen in IncFIA, IncN, and IncP plasmids and is also consistent with the structure of replicons from other groups, such as IncHI1 and IncY, although in the case of IncN and IncY, the iterons that overlap with the Rep promoter are not inverted (43, 138). Rep dimers can also bridge Rep-bound iteron arrays, one of the proposed mechanisms of handcuffing (see panel C3). (C) Different handcuffing mechanisms. Once iteron-bound Rep arrays form, they can couple two different plasmids, a reaction in trans known as plasmid handcuffing that blocks ori melting by steric hindrance (139). Three mechanisms that pair different plasmids through iteron-bound replication initiation proteins have been proposed (43). From left to right: direct dimerization of the iteron-bound initiators (proposed for the plasmid RK6 [140]), direct interaction between arrays of iteron-bound monomers, associated with a Rep conformational change induced by iteron binding (proposed for the plasmid pPS10 [141]), and bridging via dimer formation (proposed for RK2 [139]). Chaperones counteract handcuffing by facilitating the dissociation of dimers to monomers or by increasing monomer-to-dimer ratios (139, 142).
