Figure 6.
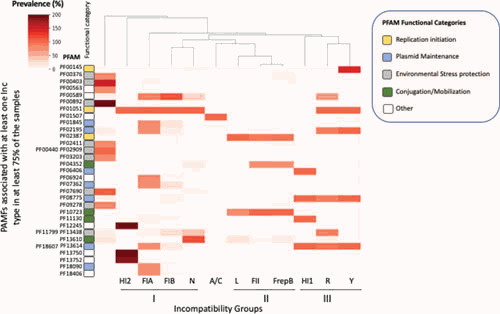
PFAM representation in the vicinity of plasmid replicons. Completely assembled E. coli plasmid sequences obtained from NCBI as of April 2019 were grouped by Inc type based on the diagnostic PCR amplicons for the PBRT system (82). Within each incompatibility group, we looked for PFAM matches within a 10-kb window centered around the diagnostic PCR amplicons. PFAM protein families present in at least 75% of the samples in at least one Inc group were identified and mapped to individual open reading frames. These PFAMs were also identified in other Inc groups. These PFAM representation data were compiled into a matrix and plotted using color-coding, with darker colors representing a higher prevalence of protein families within each Inc group. Inc groups are arranged by similarity using a hierarchical clustering algorithm for PFAMs. Note that the percentage can be higher than 100% in the case of duplications or if two domains are found in the same ORF. PFAMs are listed in the y axis, grouped in the following color-coded functional categories. Replication initiation-related PFAMs (orange): m5C methylase (PF00145), initiator replication protein (PF01051), and IncFII RepA protein family (PF02387). Plasmid maintenance PFAMs (light blue): PemK endoribonuclease toxin (PF01845) ParB-like nuclease domain (PF02195), StnA protein (PF06406), post-segregation TA CcdA (PF07362), ParB family (PF08775), ParA AAA+ and HT domains (PF13614 PF18607), centromere-binding protein HTH domain (PF18090). Environmental stress protection PFAMs (light gray): MerR family regulatory protein (PF00376), heavy metal-associated domain (PF00403), EamA-like transporter family (PF00892), mercuric transport protein (PF02411), tetracycline repressor family (PF02909 PF00440), MerC mercury resistance protein (PF03203), major facilitator superfamily (PF07690), MerR (PF09278), impB/mucB/samB Y family polymerase (PF13438, PF11799). Conjugation/mobilization PFAMs (green): phage integrase family (PF00589), ProQ/FINO family (PF04352), replication regulatory protein repB (PF10723), TraC_F_IV F pilus assembly (PF11130), DDE domain found in transposases (PF13610). Other PFAMs (white): EAL domain (PF00563), phosphoadenosine phosphosulfate reductase family (PF01507), DUF1281 (PF06924), bacterial IgE domain (PF12245), bacterial IgE-like domain (group3) (PF13750), DUF4165 (PF13752), ferredoxin-like domain in Api92-like protein (PF18406).
