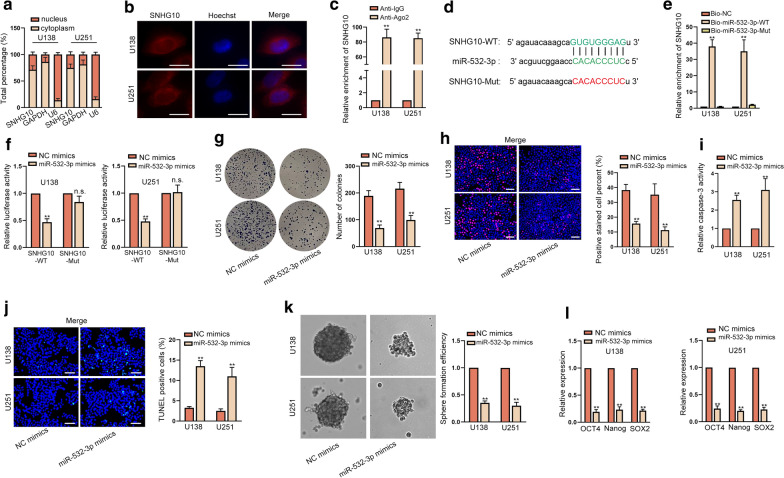
Fig. 3.
MiR-532-3p was sponged by SNHG10 and hampered cell growth and stemness in glioma. a, b Subcellular fractionation and FISH assays (scale bar = 10 μm) were performed to judge the subcellular localization of SNHG10 in U138 and U251 cells. c RIP assays demonstrated that SNHG10 could be enriched in the beads conjugated with Ago2 antibody. d Binding sites between SNHG10 and miR-532-3p were shown. e RNA pull down assay validated that SNHG10 could bind to miR-532-3p. f Luciferase reporter assay certified the impact of miR-532-3p on the luciferase activity of SNHG10-WT/Mut. g, h The proliferation of cells with or without miR-532-3p elevation was examined by colony formation and EdU assays (scale bar = 100 μm). i, j Cell apoptosis was probed by caspase-3 activity detection and TUNEL assay (scale bar = 100 μm) upon miR-532-3p overexpression. k, l Sphere formation assay and RT-qPCR examined the effect of overexpressed miR-532-3p on stemness. **P < 0.01, n.s. presented no significance