Fig. 3. Decoding polymer sequences using aerolysin pores and deep learning.
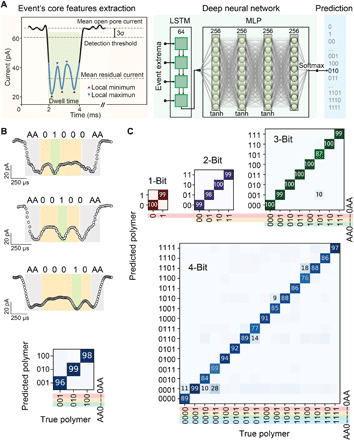
(A) Details for nanopore signal processing and relative deep learning workflow. (B) Characteristic translocation events of polymers containing bit-1 at different positions, i.e., A01000AA, AA00100AA, and AA00010AA, and the corresponding confusion matrix results obtained by deep learning. (C) Confusion matrix of 1-, 2-, 3-, and 4-bit polymer sequences classification shown for a selection percentage of 10% (see figs. S35 to S38 for dependence on selection threshold). Columns represent true polymers from the test set, while rows are the polymers that deep learning assigned them to. All data were obtained using 1.0 M KCl, 10 mM tris, and 1.0 mM EDTA buffer at pH 7.4, applying a bias potential of 100 mV.
