Fig. 4. SARS-CoV-2 Mpro in complex with GC-376 analogs.
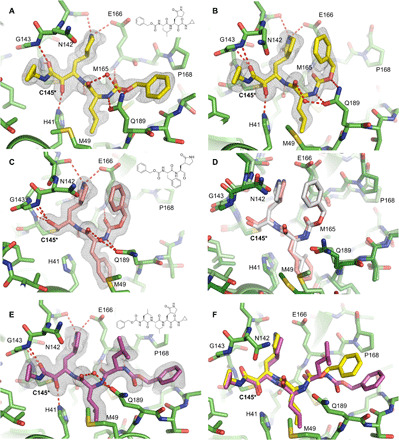
Unbiased Fo-Fc electron density map, shown in gray, is contoured at 2 σ. Hydrogen bonds are shown as red dashed lines. Solved as a dimer in the P21 spacegroup, two different conformations of the Cbz group of UAWJ246 were observed in the (A) protomer A and (B) protomer B. (C) The complex structure of UAWJ247, revealing that the P2 position can accommodate a Phe side chain. (D) Comparison of the binding poses of UAWJ247 (dark green/salmon) and GC-376 (light green/gray, PDB ID: 7BRR). (E) The complex structure of UAWJ248, solved as a dimer in the P1 space-group. Protomer A is shown here, and the inhibitor binding pose is identical in protomer B (fig. S8). (F) Comparison of the binding poses of UAWJ248 (purple) and UAWJ246 (yellow) in protomer A.
