Fig. 5. Single-cell gene expression analyses of PBMCs from COVID-19–infected, influenza-infected, and healthy participants demonstrate profound differences in the relative abundance and transcriptional activity of cell subsets across conditions.
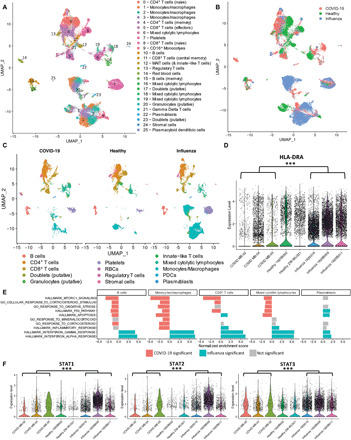
(A) Uniform Manifold Approximation and Projection (UMAP) plots depict transcriptional clusters, which (B) vary transcriptionally as a function of condition despite the presence of nearly all subsets across various conditions, as evidenced in (C). (D) Violin plots demonstrate significant down-regulation of HLA-DRA among all cells from COVID-19–infected patients compared to influenza-infected patients (with healthy controls included for reference). Asterisks indicate significance at Bonferroni-corrected P values < 0.001. (E) GSEA analysis of gene expression differences between COVID-19 and influenza groups across major cell subsets. In direct comparison to cells from influenza-infected patients, transcriptional patterns among cells from COVID-19–infected patients reveal significant up-regulation (red bars) of metabolic pathways, stress pathways, and glucocorticoid signaling pathways across major cell subsets, particularly monocytes/macrophages. In contrast, interferon pathways were significantly down-regulated (blue bars) among subsets from COVID-19–infected patients compared to those from influenza-infected patients. Gray bars indicate that tests for enrichment did not meet statistical significance for a particular subset. (F) Violin plots demonstrate significant down-regulation of STAT1, STAT2, and STAT3 among monocytes/macrophages from COVID-19–infected patients compared to influenza-infected patients (with healthy controls included for reference). Asterisks (***) indicate significance at Bonferroni-corrected P values of <0.001.
