Fig. 5. Measurements of residual disease.
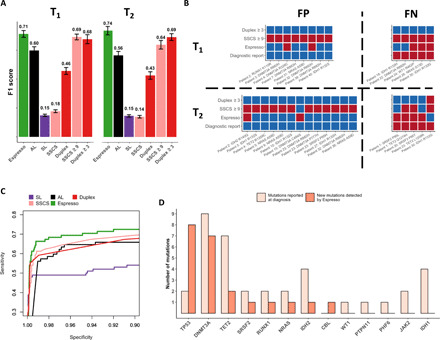
(A) Espresso provides a preferred balance between precision (PPV) and recall (sensitivity), as determined by the inspection of 78 SNVs reported across 35 of 42 patients at the time of AML diagnosis. Mutations were called in the patients’ sample at 21 different iterations. In each iteration, 6 random patients of the 42 were excluded. Median F1 scores and 1 SD are shown for the various methods tested at two time points during the course of treatment (T1 and T2, Wilcoxon signed-rank test: P ≤ 6.4 × 10–5 for all the comparisons with Espresso). (B) The variation in the mutations being called by Espresso (α ≤ 0.05, Bonferroni-adjusted), SSCS (≥9 nonreference supporting reads), and duplex (≥3 nonreference supporting reads) is illustrated. Red color indicates called mutations, while blue color indicates that mutations were not detected. FP, false positives; FN, false negatives. (C) Sensitivity versus specificity as determined by the different tested methods. (D) Enrichment of clones, carriers of TP53, and DNMT3A mutations is observed in patients with AML following therapy. The y axis represents the number of mutations detected, classified by the affected genes.
