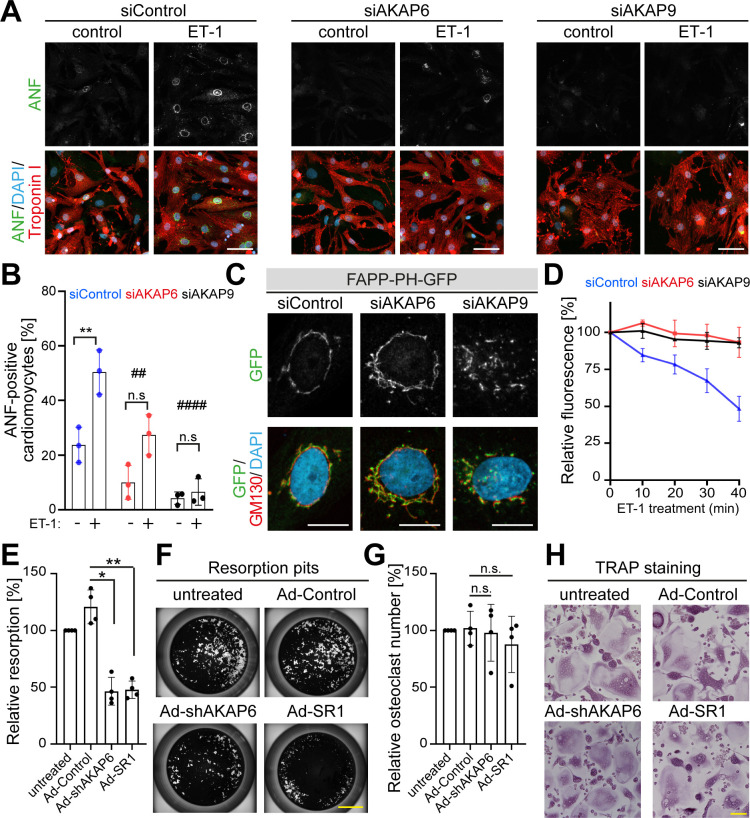
Figure 9. AKAP6 and AKAP9 dependent Golgi localization to the nuclear envelope is required for cell-specific functions in cardiomyocytes and osteoclasts.
(A) Immunostaining of ANF (green), troponin I (red), and DNA (DAPI) of P3 cardiomyocytes transfected with siControl, siAKAP6, or siAKAP9 and stimulated with vehicle or 100 nM ET-1 for 24 hr. Cells were analyzed for a hypertrophic response (perinuclear ANF expression). (B) Quantitative analysis of the percentage of ANF-positive troponin I-positive cardiomyocytes. Data are represented as individual biological replicates, together with mean ± SD from three independent experiments. Statistical analysis was performed with two-way ANOVA with post-hoc Bonferroni comparison. *: p<0.05, **: p<0.01, ****: p<0.0001, n.s.: no significance; ##: p<0.01, ####: p<0.0001 compared to siControl + ET-1. (C) Immunostaining of GM130 and DNA (DAPI) in control, AKAP6- and AKAP9-depleted cardiomyocytes transfected with FAPP-PH-GFP indicating that depletion of AKAP6 and AKAP9 displaced most of the FAPP-PH-GFP signal from the nuclear envelope. Scale bars: 10 µm. (D) Quantitative analysis of C. Individual regions of GFP fluorescence in siControl-, AKAP6- and AKAP9-treated P3 cardiomyocytes transfected with FAPP-PH-GFP were monitored with confocal microscopy after treatment with vehicle or 100 nM ET-1 and followed for 40 min. Relative fluorescence of GFP was normalized to time 0. 15 cells per condition were measured and pooled from three independent experiments. Error bars represent the SD. (E) Relative resorbed area of osteoclasts transduced with the indicated adenovirus for the last 3 days of differentiation in calcium phosphate-coated wells. Data are ± SD of 4 independent experiments. Statistical analysis was performed with One-way ANOVA with Bonferroni comparison. *: p=0.015, **: p<0.01. (F) Representative micro images. Scale bars: 500 μm. (G) Relative osteoclast number after transduction of the indicated virus for the last 3 days of differentiation. TRAP-positive cells with ≥3 nuclei were considered as osteoclasts. Data are mean ± SD of 4 independent experiments. Statistical analysis was performed with one-way ANOVA with Bonferroni comparison. n.s.: no significance. (H) Representative images. Scale bars: 100 μm.