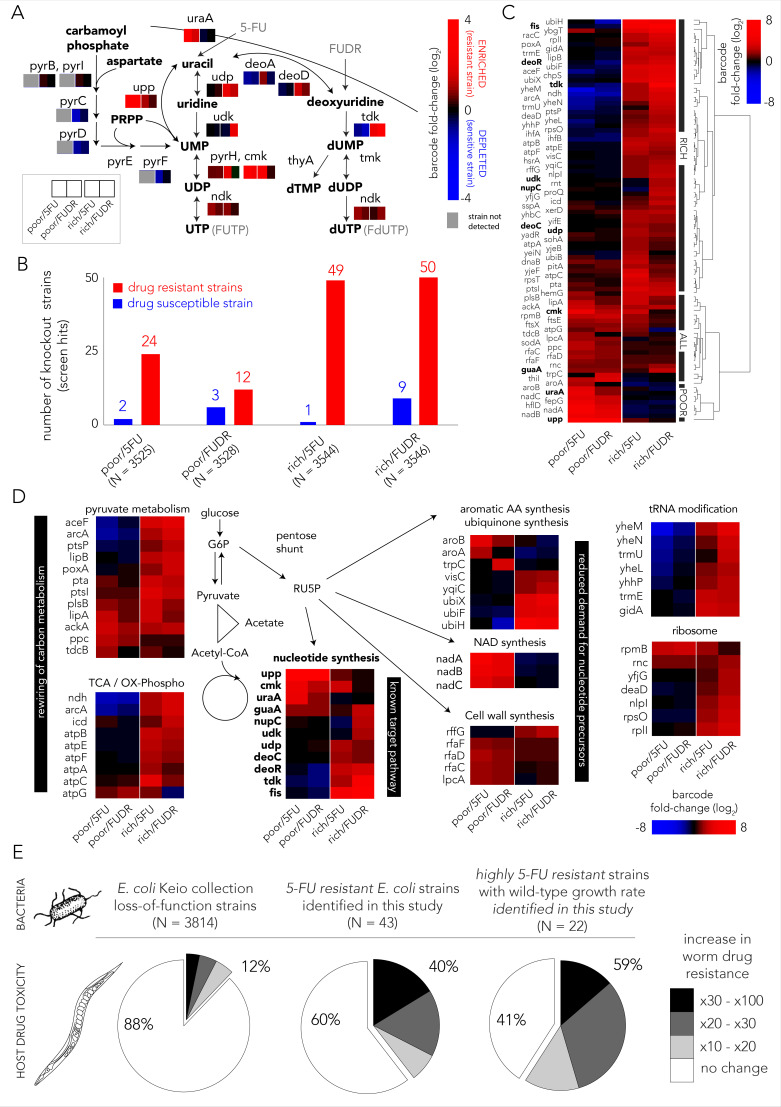
Figure 2. Multiple gene knockouts increase drug resistance in bacteria and are expected to lower drug toxicity in the Caenorhabditis elegans host.
(A) Screen results projected on the network of nucleotide synthesis. Compatible with the known toxicity mechanism, we observed that mutations in the salvage and pro-drug activation pathways increase drug resistance (red) while loss-of-function mutations in the de-novo synthesis pathway increase drug sensitivity (blue). (B) The number of strains found to be differentially represented in our screens. Hits were determined by fold-change and false-discovery-rate adjusted p-value that was calculated with DEBRA and DESeq2 tools. The number of hits in nutrient-rich media is considerably higher than the number of hits in nutrient-poor media. (C) Unsupervised hierarchical clustering of hits identified in all screen conditions. The clustering uncovers a media-dependent Resistome pattern. The majority of hits are nutrient-rich media specific. Only a minority of hits are nutrient-poor media specific. The genes from nucleotide synthesis network are marked bold. (D) The functional characterization of identified hits reveals a plausible metabolic connection through RU5P, a metabolite from the pentose phosphate pathway that is a precursor for nucleotide synthesis. Hits were assigned to broad functional categories according to their annotated function in the KEGG and EcoCyc databases. (E) Gene-knockouts that increase bacterial resistance will likely reduce drug toxicity in the C. elegans host. The three pie charts show the proportion of bacterial strains that reduce drug toxicity in C. elegans for different subsets of bacterial strains: all tested strains from the Keio library collection (left) previously tested in the C. elegans screen (Scott et al., 2017), all 5-FU resistant bacterial strains we identified in our screens and that are present in the C. elegans screen (middle), and a subset of highly 5-FU resistant strains we identified (fold enrichment > 6) that also have an un-impaired growth rate in either nutrient-rich or nutrient-poor media (right).