Figure 1.
Functional Diversification of SRPK to Control Developmental Ubiquitin Signaling
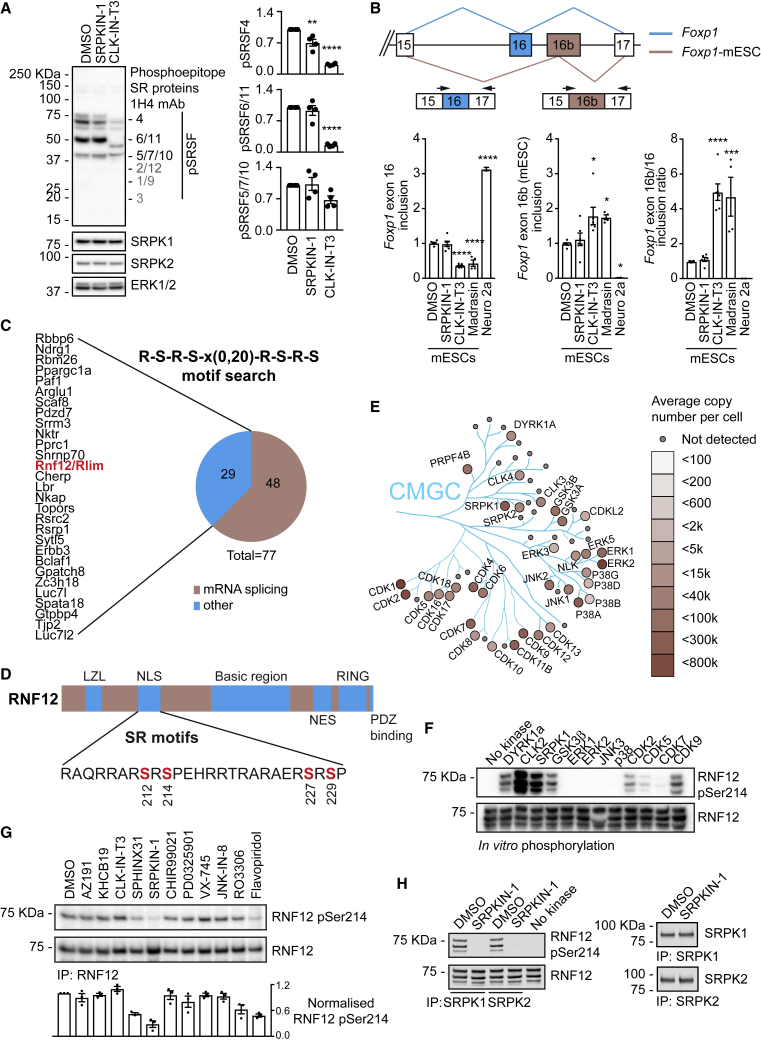
(A) Wild-type (WT) mESCs were treated with 10 μM SRPKIN-1 or CLK-IN-T3 for 4 h, and phosphorylation of Ser-Arg rich splicing factors (SRSF) was assessed (Left). SRSF phosphorylation, SRPK1, SRPK2, and ERK1/2 levels were determined by immunoblotting. Expected positions of SRSFs that are not detected are shown in gray. Quantification of SRSF phosphorylation (Right). Data represented as mean ± SEM (n = 4). One-way ANOVA followed by Tukey’s multiple comparisons test; confidence level 95%. pSRSF4: (∗∗) p = 0.0032, (∗∗∗∗) p < 0.0001, pSRSF6/11: (∗∗∗∗) p < 0.0001.
(B) Splice variants of Foxp1 mRNA including mutually exclusive exons 16 (Foxp1, GenBank: NM_053202.2, cyan) or 16b (Foxp1-ESC, GenBank: XM_030255074.1, tan) (Top). mESCs were treated with 1 μM SRPKIN-1 or CLK-IN-T3, or 10 μM Madrasin for 8 h, and Foxp1 exon 16-16b incorporation determined using specific quantitative RT-PCR primers. Neuro 2a is a control for exon 16b exclusion in differentiated cells (Bottom). Data represented as mean ± SEM (n = 3). One-way ANOVA followed by Tukey’s multiple comparisons test; confidence level 95%. Exon 16 inclusion: (∗∗∗∗) p < 0.0001, Exon 16b inclusion: (∗) p = 0.0164, p = 0.0485, and p = 0.0489 (left to right). Ratio exon 16b/16: (∗∗∗∗) p < 0.0001, (∗∗∗) p = 0.0003.
(C) SRPK substrates predicted using ScanProsite and grouped according to UniProt functions.
(D) RNF12 phosphorylation sites detected by mass-spectrometry. LZL, leucine-zipper like; NLS, nuclear localization signal; NES, nuclear export signal; RING, RING E3 ubiquitin ligase catalytic domain.
(E) CMGC family kinase copy numbers in mESCs determined by quantitative proteomics and represented using Kinoviewer.
(F) CMGC kinase (200 mU) phosphorylation of the RNF12 SR-motif in vitro was determined by immunoblotting for RNF12 phospho-Ser214 and total RNF12.
(G) mESCs were treated with 10 μM of the following kinase inhibitors: AZ-191 (DYRK1B), KH-CB19 (CLK-DYRK), CLK-IN-T3 (CLK), SPHINX31 (SRPK1), SRPKIN-1 (pan-SRPK), CHIR-99021 (GSK-3), PD-0325901 (MEK1/2), VX-745 (p38), JNK-IN-8 (JNK), RO-3306 (CDK1), and flavopiridol (CDK7/9) for 4 h and RNF12 SR-motif phosphorylation determined by immunoblotting for RNF12 phospho-Ser214 and total RNF12. Normalized RNF12 Ser214 phosphorylation is shown below. Data represented as mean ± SEM (n = 3).
(H) SRPKIN-1 inhibition of SRPKs in vivo was determined by pre-treatment of mESCs with 10 μM SRPKIN-1 for 4 h followed by SRPK1 or SRPK2 immunoprecipitation kinase assay using RNF12 as a substrate. RNF12 SR-motif phosphorylation was analyzed by immunoblotting for RNF12 phospho-Ser214 and RNF12. SRPK1 and SRPK2 levels are shown as a loading control, Related to Figure S1; Tables S1 and S2.