Figure 1.
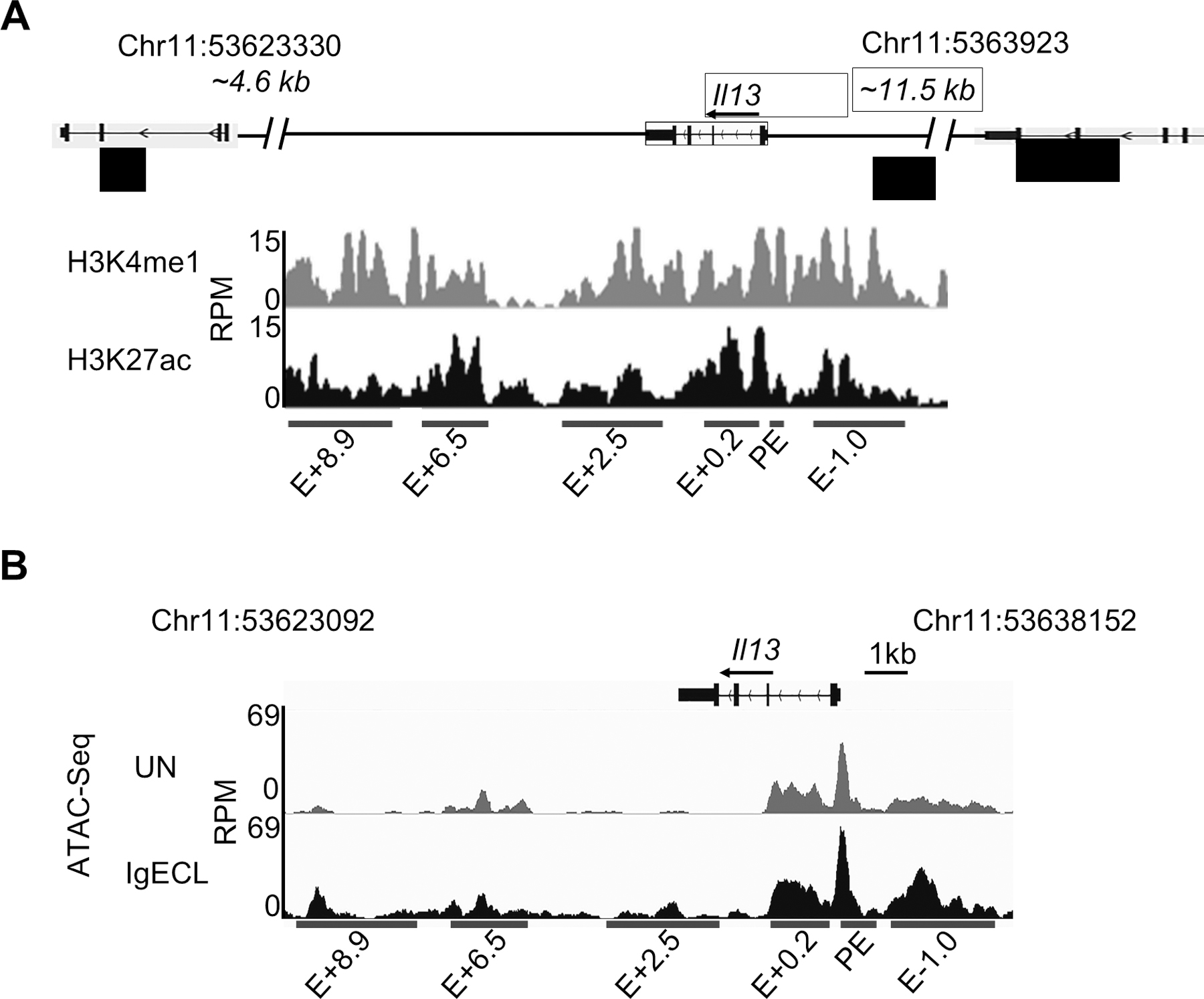
Identification of putative Il13 enhancers. (A) H3K27ac and H3K4me1 ChIP-Seq tracks from chromosome position 53623092 to 53630153 (7,061 bp), which is located in the DNA regions between the Il13 and Il4 gene (4.6 kb to the TSS of the Il4 gene) and between the Il13 gene and Rad50 gene (11.5 kb to TSS of the Il13 gene) in resting BMMCs. Red bars indicate putative Il13 enhancers and promoter positions which showed significant H3K4me1 and H3K27ac modifications. E: enhancer; +0.2, +2.5, +6.3, +8.9 indicates the distance (kilo base pair) from the beginning of the downstream Il13 enhancer to the TSS of the Il13 gene, and −1.0 indicates the distance from the beginning of the upstream Il13 enhancer to the TSS of the Il13 gene. RPM: reads per million. (B) Omni-ATAC-seq tracks at the Il13 gene obtained from BMMCs that were not stimulated (UN) or stimulated with IgE receptor crosslinking (IgECL). Data represent two biological samples.
