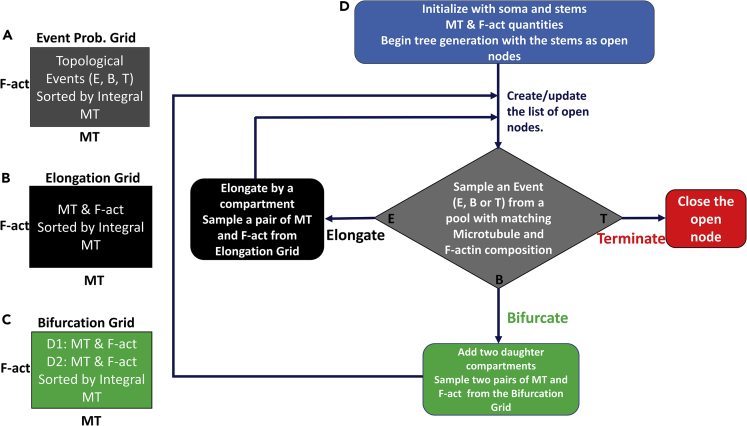
Figure 4.
Description of the Generative Model of MT- and F-act-Driven Virtual Dendritic Arbor Generation
(A–D) Stochastic sampling is carried out from three data grids (A–C) binned by local microtubule (MT) and F-actin (F-act) quantities. The grids are used to sample (A) branch topology (elongation, bifurcation, or termination), as well as the cytoskeletal compositions for the child compartments of elongations (B) and bifurcations (C). All data are sampled from the bin that best matches the MT and F-act composition of the active compartment, with a bias for higher integral microtubule values (see Methods for definition). The tree generation process (D) starts with the initial MT and F-act concentration of the soma and the dendritic stem as active compartments. For each active compartment, branch topology sampling determines whether to elongate (adding one new compartment), bifurcate (adding two new compartments), or terminate (ending the branch). If a node elongates or bifurcates, the cytoskeletal composition of the newly created compartment(s) is sampled from the corresponding data grids. The process is repeated for all newly added nodes, until all branches terminate. The model described here is employed for all three neurons types in this study.