Figure 3.
ParB foci Do Not Disassemble for a Long Time Before Splitting Events
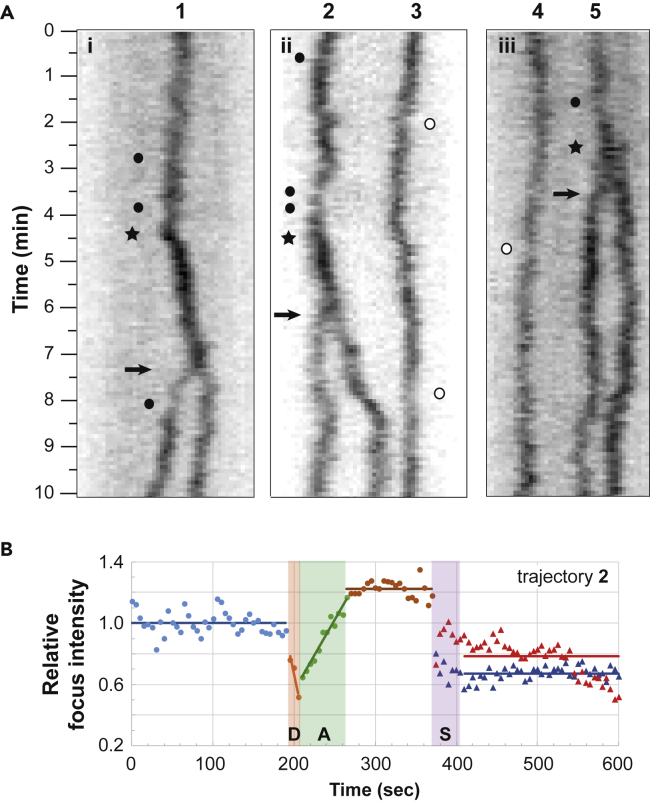
(A) The trajectories of the ParBF-mTq2 foci, labeled 1 to 5, positioned over the nucleoid length, were obtained by kymographs analyses. E. coli cells (i-iii), carrying a mini-F plasmid (pJYB249) with its endogenous ParABSF system, expressing a ParBF-mTq2 functional fusion protein, were observed by time-lapse epi-fluorescence microscopy. Images were collected every 5 s over 10 min and converted to kymograph (see Transparent Methods). The black arrows indicate the splitting events of ParB foci. Note that the ParB foci intensity increases strongly (black star) 1.5 to 3 min before the splitting events. Closed and open circles indicate low ParB fluorescence intensities in traces with or without splitting, respectively. The panel width corresponds to the length of the nucleoid.
(B) Intensity of ParBF fluorescent foci before and after the splitting event. The integrated ParBF-mTq2 fluorescence (thin line) from trajectory number 2 in (A) is measured every frame and plotted over time with color data points. The signal was subtracted from the average background level and normalized to 1 from the average intensity before the drop in intensity (blue dots). Dots and triangles represent the intensity of one and two ParB foci before and after the splitting event (S), respectively. R and L represent the putative centromere replication and the ParB loading steps, respectively. The blue, brown, red, and blue horizontal bars represent the mean values of the normalized ParB foci intensity of the one focus before R, before S, and the 2 foci after S, respectively. The orange and green bars represent the linear regression of the variation in ParB fluorescent intensities. Note that replication (R) is inferred from the increase in foci intensity that (I) occurs between the 2 plateau of ParB fluorescence mean intensity and (ii) takes place less than 5 min before splitting (S).
See also Figure S3.