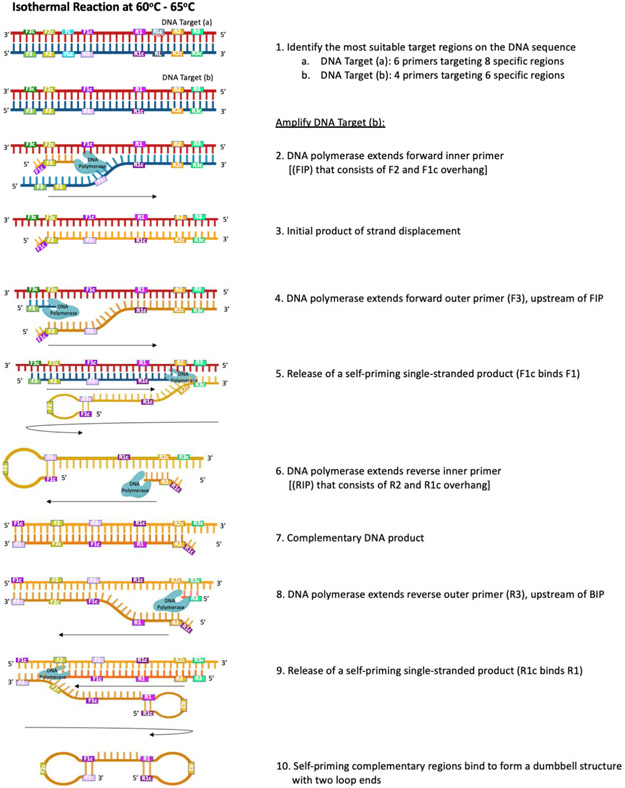
Figure 4.
Molecular underpinnings of a loop-mediated isothermal amplification (LAMP) reaction (shown with DNA template). [1] Six – eight target regions are included in a LAMP assay; [1a] 6 primers (including forward and reverse loop primers) target 8 regions to provide more sites for priming and increase the rate of amplification, as this affords an increase in concatemer formation, while [1b] 4 primers target 6 regions in a standard reaction. [2] The reaction begins as the forward inner primer (FIP), which contains a sequence for the F2 region of target DNA and a complementary sequence overhang (F1c), binds the F2c region on the 3’ end of the target. A strand-displacing DNA polymerase (e.g., Bst DNA polymerase) extends the FIP to [3] synthesize a complementary strand of DNA (the initial product). Next, [4] forward outer primer (F3) binds to a region upstream of the FIP binding site; it is extended by DNA polymerase to displace the initial product, which [5] self-primes into a single-end loop structure as F1c binds F1. [6] The reverse inner primer (RIP), which contains a sequence for the R2 region of the target DNA and a complementary sequence overhang (R1c), binds the R2 region on the 3’ end of the initial product; [7] DNA polymerase synthesizes complementary DNA in the 3’ to 5’ direction. [8] Reverse outer primer (R3) binds to a region upstream of the RIP binding site; it is extended by DNA polymerase to [9] displace the self-priming dumbbell product with two loop ends, which forms as [10] F1 binds F1c and R1c binds R1. This dumbbell structure forms the base for the cycling phase of the LAMP reaction, wherein multiple sites are available for binding by primers (FIP, BIP, and optional loop primers); these sites initiate the synthesis of concatemers (not pictured) that also contain initiation sites, which are subsequently used during the cycling phase, increasing overall amplification.