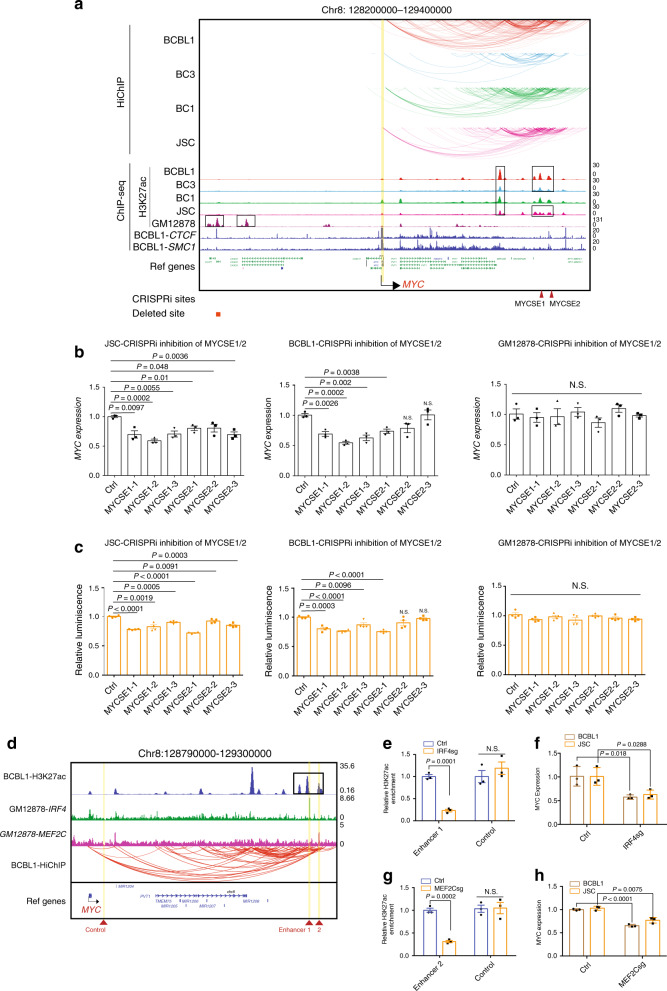
Fig. 2. MYC SEs control MYC expression and are essential for PEL cell growth.
a PEL H3K27ac HiChIP and ChIP-seq tracks, GM12878 H3K27ac, BCBL1 CTCF, and SMC1 ChIP-seq tracks are shown. Black arrow and vertical yellow line indicate MYC. Curved line indicates significant HiChIP link. Black boxes indicate SEs. The positions of CRISPRi sgRNAs (MYCSE indicates MYC SE) or dual gRNAs deletion are indicated at the bottom. b BCBL1, JSC, or GM12878 cells stably expressing dCAS9-KRAB-MeCP2 were transduced with sgRNA targeting the MYC SE or non-targeting control. qRT-PCR was used to quantitate the MYC expression. The levels of control sgRNA-treated cells was set at 1 (n = 3, independent experiments). c Cell growth following CRISPRi treatment was determined by CellTiter Glo luminescent assay that measures live cell number (n = 4, independent experiments). d BCBL1 H3K27ac, GM12878 IRF4, MEF2C ChIP-seq, and BCBL1 HiChIP (red curved lines) tracks at the MYC locus. MYC is indicated by black arrow. MYC SE is indicated by black box. Enhancer 1 indicates site of GM12878 IRF4 ChIP-seq peak that overlapped with MYC SE and enhancer 2 indicates site of GM12878 MEF2C ChIP-seq peak that overlapped with MYC SE. Genomic site near MYC promoter that lacked H3K27ac signals was used as control. Yellow lines indicate the positions for qPCR primers used in ChIP assays. e IRF4 expression was knock out using CRISPR in BCBL1 cells. SE H3K27ac signals were determined by ChIP-qPCR following CRISPR knock out. The levels of non-targeting control sgRNA-treated cells were set to 1 (n = 3, independent experiments). f MYC expression in BCBL1 and JSC cells following IRF4 knock out. qRT-PCR was used to determine the MYC expression levels. The levels of control sgRNA-treated cells were set to 1 (n = 3, independent experiments). g MEF2C expression was knocked out using CRISPR in BCBL1 cells. SE H3K27ac signals were determined by ChIP-qPCR following CRISPR knock out. The levels of non-targeting control sgRNA-treated cells were set to 1 (n = 3, independent experiments). h MYC expression in BCBL1 and JSC cells following MEF2C knock out. qRT-PCR was used to determine the MYC expression levels. The levels of control sgRNA-treated cells were set to 1 (n = 3, independent experiments). A two-tailed unpaired t-test was used for all statistical analyses. The error bars indicate the SEM for the averages across the all experiments. Source data are provided as a Source Data file.