Abstract
The Vibrio genus inhabit estuarine and marine ecosystem throughout the world and can cause severe infections in humans and animals. Previous studies have demonstrated the dynamics of Vibrio at both community and population levels and assessed the close relationship between environmental factors and Vibrio diversity and abundance, such as temperature, salinity, and dissolved oxygen. It is also generally believed that aquaculture is the fastest-growing food sector, which is also applying great environmental impacts on microbial communities in aquatic ecosystems. However, our understanding of the spatiotemporal quantification of Vibrio throughout the four seasons in the aquaculture zone and response to environmental factors remains poor. To explore the spatiotemporal distribution and abundance of the Vibrio community with their related environmental factors and detect the relationships among them, we collected 10 seawater sites spanning four seasons across the whole year in Dongshan Bay for investigating the Vibrio community dynamics. Marked differences in diversity and abundance of the Vibrio community were observed between seasons, which were mainly driven by temperature, dissolved oxygen, nitrate, and nitrite. qPCR analysis showed that Vibrio abundance was most abundant in the summer (5.37 × 106 copies/L), compared with the autumn (4.58 × 106 copies/L), spring (1.18 × 106 copies/L), and winter (1.55 × 104 copies/L). A total of 22 Vibrio operational taxonomic units (OTUs) and 28 species were identified by universal bacteria 16S rRNA gene and cultivation methods, with Vibrio fortis the dominant in these aquaculture areas. To summarize, our present study is one of the few studies to research the occurrence of Vibrio in marine aquaculture of South China, and the results indicate that Vibrio are widely distributed in aquaculture environment and that a further risk assessment is needed to be conducted.
Keywords: seasonal pattern, aquaculture activity, seawater, Vibrio diversity, ecological distribution
Introduction
Currently, more than 47% of the global fish production are harvested from aquaculture, and China is the world’s largest producer of aquaculture-based foods, representing ca. 60% of global production; and marine aquaculture accounts for about 6% of our total domestic food supply (Food and Agriculture Organization of the United Nations [FAO], and The State of World Fisheries and Aquaculture [WFA], 2016). The substantial supply of nutrients can modify the local physicochemical environment and alter the microbial community structure in the aquaculture area (Dowle et al., 2015). The total bacterial populations mediate crucial biogeochemical processes in aquatic ecosystems that have been studied for a long time (Hall et al., 2008; Fonte et al., 2013; Zhu et al., 2019). However, seasonal changes affect the microbial community composition, and biomass is often attributed to responses to changes in environmental conditions driven by seasonal climate cycles (Ward et al., 2017).
The genus Vibrio is a group of gram-negative rods belonging to the Gammaproteobacteria with facultative fermentative metabolisms. This genus is highly heterogeneous and has abundant members of native microbial assemblages in a great variety of aquatic environment (Thompson et al., 2004; Zhang X.H. et al., 2018). More than 130 Vibrio species have been described to date; several Vibrio spp. are well known as a causative agent of foodborne-related illnesses (e.g., foodborne gastroenteritis), which can have a huge impact on public health and management (Pruzzo et al., 2005; Lee and Raghunath, 2018; Zhang X.H. et al., 2018). Most earlier studies on the Vibrio diversity are carried out by cultivation-dependent approaches (Baffone et al., 2006; Hsieh et al., 2008; Nigro et al., 2011; Amin et al., 2016; Kopprio et al., 2017; Wong et al., 2019). Previous studies estimated that the specificity of thiosulfate citrate bile salt sucrose (TCBS) medium for the genus Vibrio was approximately 60% (Oberbeckmann et al., 2012). Due to the rapid development of high-throughput sequencing (HTS) technology, it offers potential solution to overcome the limitation of traditional cultivation-dependent approach.
The ecological importance of Vibrio has drawn increasing attention in the study of the Vibrio dynamics in estuarine and coastal habitats worldwide (Thompson et al., 2004; Mansergh and Zehr, 2014; Lopez-Hernandez et al., 2015; Vezzulli et al., 2016; Liang et al., 2019). At a global scale, the abundance of Vibrio has been correlated with temperature and salinity in most marine environments (Thompson et al., 2004; Julie et al., 2010; Nigro et al., 2011; Oberbeckmann et al., 2012; Vezzulli et al., 2016; Zhang X.H. et al., 2018). Moreover, inorganic nutrients, such as dissolved organic carbon (DOC), dissolved inorganic carbon (DIC), NO3–, NO2–, and PO43–, which can be directly utilized by Vibrio for metabolism, also exert significant influences on Vibrio abundance, motility, and community composition (Thompson and Polz, 2006; Kopprio et al., 2017; Liang et al., 2019). Although numerous and varying environmental parameters increase the complexity and necessity of studying the ecology of Vibrio in different sea areas, comprehensive investigations are necessary to define the relationship between the environment factor and Vibrio dynamics, especially comparing the effects of seasonality and aquaculture activity on the assembly processes of Vibrio communities.
Fujian Province is one of the most important aquaculture areas in South China, and the offshore water quality there also severely impacted human activities due to industry, agriculture, and a dense human-made population. The Dongshan Bay is located in the southwest of the Taiwan Strait, south of Fujian Province, China. Importantly, in recent years, increasing coastal water pollution has emerged in Dongshan Bay, which was mostly caused by human activities including increasing discharge of aquaculture, and industrial and wastewater effluent in offshore areas, which has been addressed by a previous study (Zhang W. et al., 2018). Although we know little about the large-scale distribution patterns of Vibrio in marine aquaculture ecosystems, several human pathogenic species, including Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus, are common in marine mammals, fish, and shellfish. Especially during the warm summer months, bioaccumulation in oysters and other filter feeders transmitted to humans via contaminated water or seafood, which is one of the main causes of foodborne diseases worldwide, has been widely described (Jesser and Noble, 2018). Comparing zones and their respective Vibrio diversity and abundance provides an excellent opportunity to detect changes in Vibrio communities across both seasons and aquaculture influence. However, the current information about comprehensive spatiotemporal distributions and environmental dependencies of Vibrio communities and abundance in these aquaculture ecosystems is still limited. The primary goal of our study was to understand the effects of seasonality and their intensity of aquaculture activities on the diversity and composition of Vibrio communities, which provided a valuable opportunity to demonstrate the influences of typical seasonal change on biogeochemical cycles in such systems. To solve this problem, especially for Dongshan Bay, in this study, we used HTS and qPCR analyses of the 16S rRNA gene to compare the effects of seasonality and aquaculture activity on the diversity, composition, and assembly of Vibrio communities in Dongshan Bay. At the same time, we evaluated the key physicochemical variables that influence the distribution of the Vibrio community.
Materials and Methods
Sample Collection and Site Description
Ten different sampling sites were investigated around Dongshan Bay (∼24°N, ∼118°E; Figure 1). Firstly, we established three sample survey within three different zones at the Dongshan Bay [each zone differs in terms of aquaculture intensity, fish farm (FF), water channel of farm (FW), and control zone (CN)] and four sites in the fish farm (DY1–4). In Figure 1, DY represents the four sites of DY1–4 because they have the same latitude and longitude, three sites in the water channel of farm zone (DS1–4), and the remaining three sites in the control zone (DS5–6).
FIGURE 1.
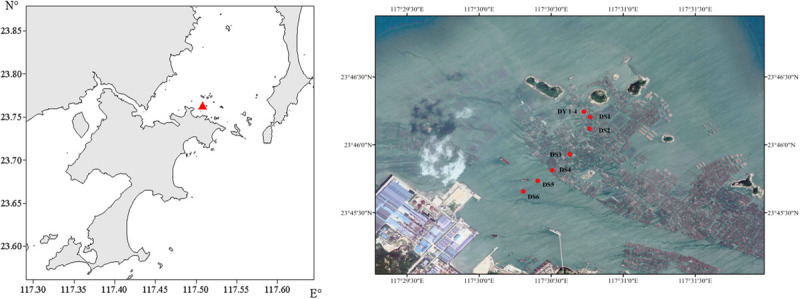
Sample area (left) and sampling stations (right).
Surface seawater at ∼1-m depth was collected with 5-L sterile bottles at the 10 sampling sites. A total of 40 water samples were collected on January 25 (winter), April 13 (spring), July 14 (summer), and October 19, 2019 (autumn) (details are given in Supplementary Table S1). About 1 L of seawater was filtered through 0.22-μm filter units (GTBP04700, Millipore), and the resulting filters were stored at −80°C until DNA extraction. Meanwhile, 500 ml of each seawater sample was collected by a sterile bottle for nutrient analysis, and the other 50 ml of water was collected in sterile vials for Vibrio isolation. Parameters such as temperature, pH, salinity, and dissolved oxygen (DO) were measured in situ with a multiparameter water quality checker (Horiba U-50, Horiba Co., Kyoto, Japan) at each sampling site at the time of sampling. The chemical indicators of water environment, including phosphate (PO43–) (standard method: GB/T12763.4–2007), nitrite (NO2–) (standard method: GB/T12763.4–2007), ammonia nitrogen (NH4+-N) (standard method: GB/T12763.4–2007), and nitrate (NO3–) (standard method: GB/T12763.4–2007), were measured following the corresponding standard methods. The environmental factors are expressed as mean ± standard deviation, and differences among groups were evaluated by one-way analysis of variance (ANOVA) using SPSS 16.0 software (IBM, SPSS, Chicago, United States).
Vibrio Isolation and 16S rRNA Identification
All the seawater samples were immediately placed in sterile coolers, then transferred to the microbial laboratory, and processed for molecular analysis within 5 h. Seawater corresponding to marine aquaculture areas was diluted with sterile seawater, and 200 μl of the appropriate decimal dilutions was plated on TCBS agar (Oxoid) both directly and after a 10-fold dilution. At room temperature (25 ± 3°C) after 48-h incubation, retrieve all the culturable heterotrophic bacteria able to grow on TCBS. From each plate, isolated colony types of each different size and morphology were picked out and streaked onto 2216E Agar to obtain pure cultures for molecular identification. The genomic DNA of each isolate was prepared following the Bacterial DNA Kit. Molecular identification at the species level was obtained by sequencing of 16S rRNA by amplifying the specific primer pair 27F (5′-TGGCTCAGATTGAACGCTGGCGG-3′) and 1492R (5′-TACCTTGTTACGACTTCACC-3′) (Hayashi et al., 2002). PCRs were performed as follows: initial denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing for 1 min at 55°C and extension at 72°C for 1.50 min, and the final elongation step at 72°C for 5 min. PCR products were sequenced using 27F/1492R primers on an ABI 3730XL automatic sequencer by Majorbio Sequencing Service (Shanghai). The sequences of all isolates was compared with that of closely related reference strains using the EzTaxon-e server. Multiple sequence alignments were performed with CLUSTAL W. The nucleotide sequence of almost the entire 16S rRNA gene of Vibrio sp. isolates was deposited at GenBank with the following accession numbers: MT269611–MT269621 (January 2019), MT269622–MT269633 (April 2019), MT269634–MT26941 (July 2019), and MT269602–MT269610 (October 2019).
DNA Extraction, PCR Amplification, and Illumina Sequencing
Genomic DNA was extracted from each filter by using the FastDNA Spin Kit for Soil and a FastPrep-24 Instrument (MP Biomedicals, United States) according to the manufacturer’s protocol. PCR amplification of V3–V4 variable regions of the 16S rRNA genes was performed using the universal primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Guo et al., 2017). Each DNA sample was PCR-amplified individually with three technical replicates. Amplicons after index added and equally pooled were then sent for the high-throughput sequencing by using Illumina HiSeq 2500 Platform (PE250) at Majorbio Bio-Pharm Technology, Shanghai, China. The raw sequencing reads have been submitted to the Sequence Read Archive (SRA) database with accession number SRR11929868–SRR11929907. Raw FASTQ files were processed and quality-filtered using the FLASH and Trimmomatic software. Operational taxonomic units (OTUs) were defined as clusters sharing 97% sequence similarity cutoff using USEARCH software (version 7.1), and chimeric sequences were identified and removed using UCHIME software (version 4.2). The Chao index and Shannon index were calculated based on OTU dilution curve analysis. The taxonomy of each representative OTU 16S rRNA gene sequence was analyzed by using the RDP Classifier (http://rdp.cme.msu.edu/) against the SILVA (SSU132) database using a confidence threshold of 70%. The quality-checked data were then analyzed on the free online platform of the Shanghai Majorbio I-Sanger Cloud Platform (www.i-sanger.com) based on QIIME 1.9.0 pipeline to obtain parameters of alpha and beta diversity indices.
Construction of Quantitative PCR Standard
Detection of the abundance of total bacteria and Vibrio spp., the primer set A-967F/B-1046R, and Vibrio specific primers 567F/680R were used to amplify the 16S rRNA gene in the aquaculture surface water samples, carried out as described in the previous study (Liang et al., 2019). Briefly, PCR standard curves were prepared from genomic DNA extracted from Escherichia coli and Vibrio alginolyticus (Marine Culture Collection of China strain, MCCC1A06468 and MCCC1A07292), which are equivalent to 102–108 gene copies per reaction. The environmental DNA and control DNA were used as DNA templates in real-time PCRs, which were carried out on LightCycler 480II Real-Time PCR system (Roche Life Science, Swiss). The real-time PCR was performed with a final volume of 25 μl, including 12.5 μl of 2 × SYBR Green Real-time PCR Master Mix, 0.2 μM for each forward and reverse primers (for bacteria) or 0.4 μM for each forward and reverse primers (for Vibrio), 2 μl of diluted (1:10) template DNA, and double-distilled water added up to 25 μl. The real-time PCR was run with the following program: 95°C for 10 min, followed by amplification (45 cycles of 15 s at 95°C and 1 min at 60°C) and a melting curve (5 s at 95°C, 60 s at 57°C or 65°C, and increasing temperature from 65°C to 98°C at 0.11°C⋅s–1 with 5 fluorescence acquisitions⋅s–1). In the qPCR analysis, each reaction was performed with three repeats, standard curves were run with every plate and ddH2O as templates were used as a no-template control (NTC). The LightCycler480 software was used for statistical analyses, and final abundance is reported as gene copies/L of seawater sampled.
Statistical Analysis
All statistical tests were considered significantly at p < 0.05. Seasonal differences in environmental parameters were evaluated using the non-parametric Mann–Whitney test. Abundances of Vibrio and bacteria were log 10(x+1) transformed before the analyses. In cases where the abundance of some samples was below the detection limit of the available method, a value of zero was assigned before logarithmic conversion. Variations in Vibrio abundance between seasons were tested for significance using the Mann–Whitney test. Spearman’s rank correlation was calculated to determine the relationship between environmental factors and Vibrio abundance. All the above analyses were performed using SPSS statistical software version 16.0.
Results
Seasonal and Spatial Variation in Environmental Variables
The environmental parameters of the seawater samples we tested, including water temperature, salinity, DO, ammonia NH4+, nitrite NO2–, nitrate NO3–, and phosphate PO43–, and detailed physicochemical parameters of water quality for 10 stations are shown in Figure 2 (Table S1). All the samples were divided into four clusters based on seasonal patterns and physicochemical parameters, which are summarized in Table 1. Water temperature and nitrite NO2– concentrations varied significantly between seasons, with temperature varying from 18.05 ± 0.31°C (mean ± standard deviation; winter) to 28.07 ± 0.22°C (summer), while and nitrite NO2– ranged from 0.07 ± 0.01 (spring) to 0.92 ± 0.04 (autumn). In addition, generally, DO decreased from spring to autumn, whereas nitrate NO3– showed opposite trends (Table 1). Our clustering analysis of the normalized environmental parameters using the average cluster method indicated that the environmental variable clustering was strongly affected by seasonal pattern, with values taken in winter and spring distinctly different from those of the other two seasons (Supplementary Figure S1).
FIGURE 2.
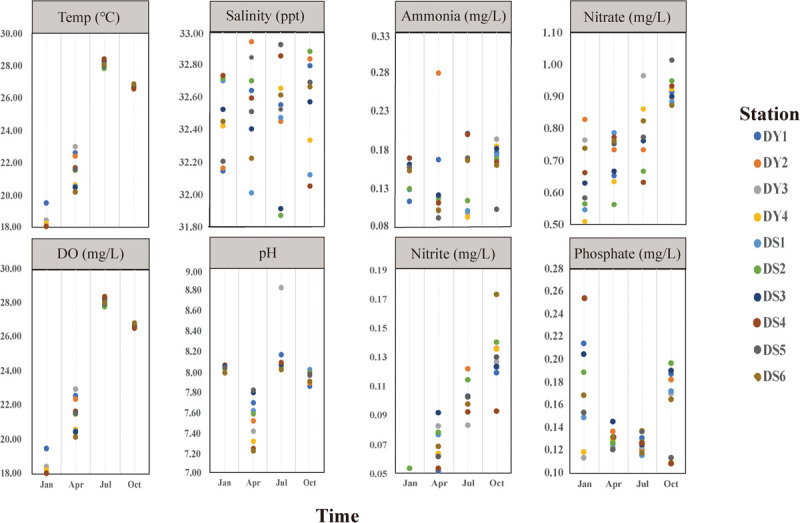
Physicochemical parameters of sampling station variables from January to October 2019 in Dongshan Bay, Fujian, China.
TABLE 1.
Summary of physicochemical parameters in four seasons.
| Physicochemical parameter |
Value |
|||
| Winter (Jan) | Spring (Apr) | Summer (Jul) | Autumn (Oct) | |
| Temp(°C) | 18.052 ± 0.315 | 21.434 ± 0.991 | 28.068 ± 0.220 | 26.675 ± 0.099 |
| pH | 8.036 ± 0.024 | 7.457 ± 0.209 | 8.123 ± 0.249 | 7.947 ± 0.052 |
| DO (mg/L) | 7.926 ± 1.020 | 7.489 ± 0.500 | 4.997 ± 0.460 | 5.153 ± 0.387 |
| Salinity(ppt) | 32.445 ± 0.226 | 32.507 ± 0.295 | 32.480 ± 0.347 | 32.561 ± 0.294 |
| NH4+(mg/L) | 0.145 ± 0.019 | 0.121 ± 0.028 | 0.132 ± 0.042 | 0.166 ± 0.008 |
| PO43–(mg/L) | 0.168 ± 0.044 | 0.130 ± 0.007 | 0.126 ± 0.007 | 0.160 ± 0.035 |
| NO3–(mg/L) | 0.665 ± 0.118 | 0.710 ± 0.076 | 0.786 ± 0.096 | 0.918 ± 0.041 |
| NO2–(mg/L) | 0.0179 ± 0.016 | 0.069 ± 0.013 | 0.101 ± 0.011 | 0.129 ± 0.020 |
Spatiotemporal Variation in Abundance of Vibrio
The Vibrio community-specific qPCR assays were performed to quantify the abundance of Vibrio in all samples, which showed an obvious seasonal pattern (Figure 3). Vibrio were most abundant across the 10 sites in summer, averaged 5.37 × 106 copies/L, significantly higher than those in winter (Mann–Whitney test, p < 0.01), with a mean value of 1.55 × 105 copies/L. Significant seasonal differences in bacterial abundance were also observed (Mann–Whitney test, p < 0.001). We compared the Vibrio abundance in different samples; the fold between the highest (JulDY4) and lowest (JanDS2) is 115. We also compared the bacteria abundance in different samples; the ratio between the highest (OctDS2) and lowest (JanDY4) is 5,614. Moreover, Vibrio also varied in abundance between different sample areas (Kruskal–Wallis, p < 0.05), the abundance of Vibrio was higher in DS1 (farm water channel) than in DS6 (control zone) (Kruskal–Wallis, p < 0.05), whereas similar variations were also observed in other seasons (Supplementary Figure S2).
FIGURE 3.
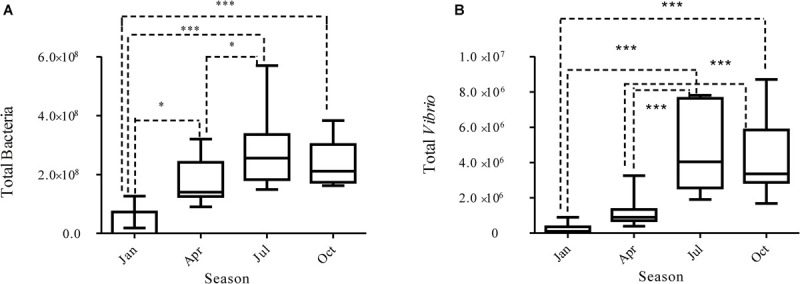
Vibrio (B) and bacterial (A) abundance determined by qPCR in different seasons.
In winter, the Vibrio abundance showed positive correlation with pH (Spearman correlation, r = 0.7354, p < 0.05) and DO (r = 0.774, p < 0.05), while only nitrate (r = 0.414, p < 0.05) showed positive correlation with Vibrio abundance in summer. Moreover, there was no significant correlation between Vibrio abundance and biogeochemical parameters in spring and autumn. Across the entire data, the abundance of Vibrio spp. not only positively correlated with temperature (r = 0.799, p < 0.01), nitrate NO3– (r = 0.672, p < 0.01), and nitrite NO2– (r = 0.771, p < 0.01) but also negatively correlated with DO (r = -0.702, p < 0.01) (Table 2).
TABLE 2.
Spearman rank correlations of qPCR results of Vibrio with environmental variables between four seasons.
| Environmental | Winter | Spring Summer | Autumn | Four seasons | ||||||||||||
| variable | r | p | N | r | p | N | r | p | N | r | p | N | r | p | N | |
| Temperature | 0.248 | 0.489 | 10 | 0.243 | 0.498 | 10 | −0.455 | 0.187 | 10 | −0.311 | 0.382 | 10 | 0.799 | 6.36E-10 | 40 | |
| pH | 0.735 | 0.015 | 10 | 0.426 | 0.220 | 10 | 0.160 | 0.659 | 10 | 0.415 | 0.233 | 10 | 0.036 | 0.827 | 40 | |
| DO | 0.774 | 0.009 | 10 | 0.170 | 0.638 | 10 | 0.152 | 0.676 | 10 | 0.201 | 0.578 | 10 | -0.702 | 4.40E-07 | 40 | |
| Salinity | −0.043 | 0.907 | 10 | -0.049 | 0.894 | 10 | 0.358 | 0.310 | 10 | 0.134 | 0.713 | 10 | 0.164 | 0.313 | 40 | |
| NH4+ | 0.246 | 0.493 | 10 | 0.267 | 0.455 | 10 | −0.588 | 0.074 | 10 | −0.188 | 0.603 | 10 | 0.077 | 0.638 | 40 | |
| PO43– | 0.365 | 0.300 | 10 | -0.226 | 0.531 | 10 | 0.018 | 0.960 | 10 | 0.541 | 0.106 | 10 | −0.119 | 0.466 | 40 | |
| NO3– | 0.309 | 0.385 | 10 | 0.0427 | 0.907 | 10 | 0.693 | 0.026 | 10 | 0.382 | 0.276 | 10 | 0.672 | 2.03E-06 | 40 | |
| NO2– | −0.067 | 0.854 | 10 | 0.128 | 0.725 | 10 | −0.432 | 0.213 | 10 | −0.109 | 0.763 | 10 | 0.771 | 5.71E-09 | 40 | |
Significant correlations (P < 0.05) were showed in bold. Correlation (r) and significance (P) values are shown.
Seasonal Succession of Vibrio Composition by 16S rRNA Sequencing
Vibrio diversity was assessed via the amplification and sequencing of the V3–V4 region of the 16S rRNA gene. In total, 1,539,256 clean reads were obtained through Illumina sequencing after quality control, ranging from 19,816 to 62,826 reads, individually. The total sequences yielded 3,057 OTUs at a 97% sequence similarity level. Of these, a total of 22,786 reads and 22 OTUs reads were selected, which were classified into the genus of Vibrio (Figure 4 and Supplementary Table S2). The mean (± SD) relative abundance of Vibrio spp. 16S rRNA gene sequence reads in water samples from summer and autumn of those stations was 0.33% (±0.12%) and 9.14% (±8.48%), respectively. The highest relative Vibrio abundance of total bacteria in the water samples (12.83%) was measured in DS6 at autumn, where station DS6 was located at the control zone.
FIGURE 4.
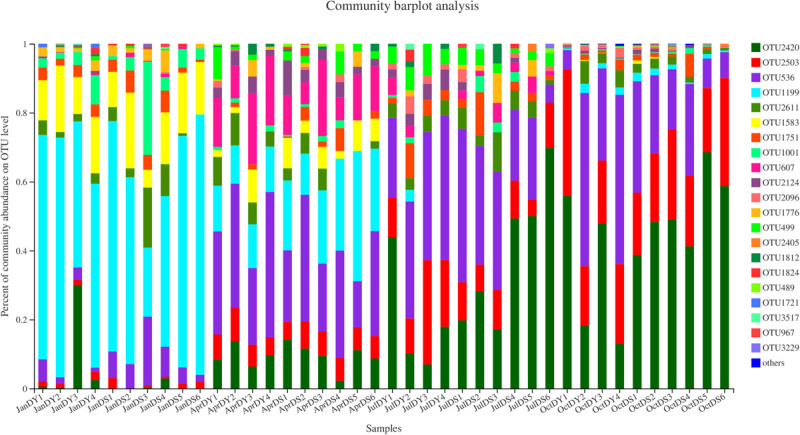
Vibrio community compositions at the species level across all samples.
Representative sequences of each OTU were compared against the EzBioCloud and National Center for Biotechnology Information (NCBI) database to determine their taxonomic status, which clustered with Vibrio aestivus, Vibrio caribbeanicus, Vibrio cortegadensis, Vibrio echinoideorum, Vibrio fortis, Vibrio furnissii, Vibrio gigantis, Vibrio hepatarius, Vibrio harveyi, Vibrio maritimus, Vibrio mimicus, Vibrio neptunius, Vibrio ponticus, Vibrio renipiscarius, Vibrio rotiferianus, Vibrio zhanjiangensis, and two unclassified Vibrio spp. (Supplementary Table S3). Our samples did not contain sequences clustering with the common pathogens Vibrio alginolyticus, Vibrio cholerae, Vibrio coralliilyticus, Vibrio parahaemolyticus, Vibrio pectenicida, Vibrio splendidus, Vibrio tubiashii, and Vibrio vulnificus. There were no significant differences (p > 0.05) in Vibrio diversity with respect to the different seasons. There are 14 OTUs detected from all the four seasons, while only one, two, and two specific OTUs were specifically found in spring, autumn, and winter samples (Supplementary Figure S3). The most dominant species was V. fortis OTU2420, occupying 44.23% of all sequences, followed by Vibrio brasiliensis OTU2503, V. harveyi OTU536, and V. gigantis OTU1199, which jointly accounted for 47.77% of all sequences (Supplementary Figure S4). OTU1812 was not resolved at the species level and showed the highest 16S rRNA gene similarity to Vibrio sp. strain r32 (NCBI Taxonomy ID AB470935) (93.01%), which may represent a novel Vibrio species.
To illustrate the diversity pattern of Vibrio spp. across all samples, the OTU-level weighted FastUnifrac analysis was conducted (Figure 5), which showed that the samples clustered together into two larger clusters and were separated into four clades according to seasonal pattern, with the exception that OctDS3, OctDY2, OctDS3, and OctDY2 are clustered with April samples showing that there is little Vibrio difference between spring and several autumn samples. In the surface water, the total average number of Vibrio OTUs in autumn (14.2 ± 2.4) is significantly higher than in summer (10 ± 2.1). The unweighted FastUnifrac analysis showed that there was no significant difference between the different terms of aquaculture intensity. After comparison of the specific Vibrio-related OTUs in different zones, it appeared that there are 20 OTUs obtained from all the three zones, while water channel samples recovered only one specific OTU (Supplementary Figure S5). These results revealed that the seasonal variations were more pronounced than the spatial variations.
FIGURE 5.
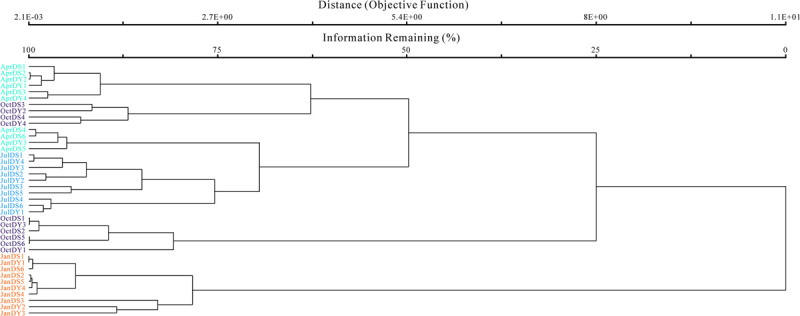
Weighted Bray–Curtis analysis was conducted at the operational taxonomic unit (OTU) level.
The relationship between environmental and spatial factors and Vibrio community components at the OTU level was investigated by the Mantel test and distance-based redundancy analyses (db-RDAs) (Figure 6). The first axis explained 18.45% of the total variance, whereas the second axis explained 10.06%. Water temperature (R2 = 0.88, p < 0.001), pH (R2 = 0.60, p < 0.001), phosphate PO43– (R2 = 0.30, p < 0.001), nitrate NO3– (R2 = 0.28, p < 0.001), and nitrite NO2– (R2 = 0.28, p < 0.001) positively correlated with RDA1, while DO (R2 = 0.89, p < 0.001) was negatively correlated with RDA1. No significant correlation was observed between salinity and NH4+ ammonia nitrogen (p > 0.05).
FIGURE 6.
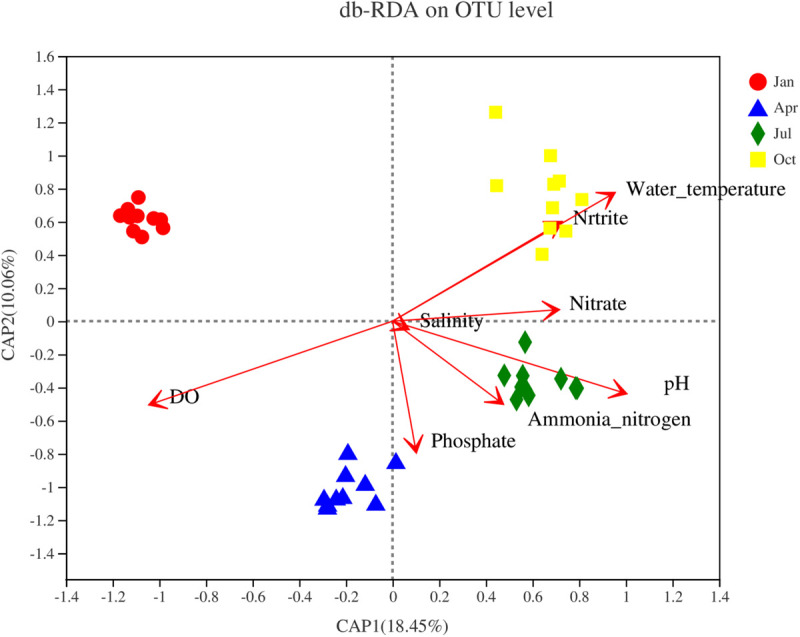
Distance-based redundancy analysis (db-RDA) illustrating the relationship between the Vibrio community at the operational taxonomic unit (OTU) level and environmental variables.
The Spearman correlations between the 22 Vibrio OTU and environmental factors were calculated and summarized in Table 3, with nine OTUs significantly correlated with four environmental factors. The most abundant species V. fortis (OTU2420) had significant correlations with DO (r = -0.602, p = 0.008), nitrate NO3– (r = -0.583, p = 0.031), and nitrite NO2– (r = -0.739, p = 0.00002). V. gigantis (OTU1199), which is the dominant group in spring, was positively related to DO (r = 0.682, p = 0.0005) and negatively related to nitrite NO2– (r = -0.573, p = 0.044) and temperature (r = -0.832, p = 0.0000000123). In addition, of the two specific Vibrio OTUs in autumn, V. fortis (OTU2405) was positively collected with nitrite NO2– (r = 0.606, p = 0.014), while V. maritimus (OTU2461) was positively collected with nitrate NO3– (r = 0.670, p = 0.0009) and nitrite NO2– (r = 0.628, p = 0.006).
TABLE 3.
Correlations between percentage composition of OTU and environmental factors.
| OTU |
Correlation coefficient1 |
|||||||
| Temperature | pH | DO | Salinity | NH4+ | PO43– | NO3– | NO2– | |
| Vibrio caribbeanicus OTU607 | ||||||||
| Vibrio fortis OTU2405 | 0.606 | |||||||
| Vibrio mimicus OTU3229 | ||||||||
| Vibrio sp. OTU1812 | ||||||||
| Vibrio maritimus OTU967 | ||||||||
| Vibrio fortis OTU2420 | −0.620 | 0.582 | 0.739 | |||||
| Vibrio zhanjiangensis OTU1721 | ||||||||
| Vibrio ponticus OTU1751 | ||||||||
| Vibrio maritimus OTU2461 | 0.670 | 0.628 | ||||||
| Vibrio natriegens OTU536 | 0.619 | |||||||
| Vibrio renipiscarius OTU1001 | −0.578 | |||||||
| Vibrio aestivus OTU1824 | ||||||||
| Vibrio echinoideorum OTU1583 | −0.836 | 0.709 | −0.659 | |||||
| Vibrio sp. OTU3517 | ||||||||
| Vibrio natriegens OTU2611 | ||||||||
| Vibrio cortegadensis OTU1776 | ||||||||
| Vibrio rotiferianus OTU2096 | 0.662 | |||||||
| Vibrio hepatarius OTU2124 | ||||||||
| Vibrio pomeroyi OTU1199 | −0.832 | 0.682 | −0.572 | |||||
| Vibrio caribbeanicus OTU499 | ||||||||
| Vibrio neptunius OTU2503 | 0.638 | 0.693 | ||||||
| Vibrio furnissii OTU489 | ||||||||
Only significant correlations (P < 0.05) are shown. −, negative; boldface, P < 0.01; lightface, P < 0.05.
Identification of Vibrio Diversity by Cultivation Method
A total of 201 presumptive Vibrio spp. were isolated; most isolates were confirmed as Vibrio spp. via the 16S rRNA sequence. There were also a significant number of isolates (50 isolates) that were identified as Photobacterium spp., Shewanella spp., Enterovibrio spp., Bacillus spp., Grimontia spp., Psychrobacter spp., and Shewanella spp. A total of 28 Vibrio species were identified; the most species-rich season was spring, with 12 species (42 isolates). The winter harbored 11 species (39 isolates) and autumn with nine species (32 isolates). The lowest species-rich season was summer, with only eight species (38 isolates). Three Vibrio spp. commonly occurring in different seasons were V. fortis, V. alginolyticus, and Vibrio hyugaensis, while the most frequently isolated Vibrio sp. was Vibrio crassostreae. Two species (V. parahaemolyticus and V. brasiliensis) were isolated in summer, whereas four species (Vibrio shilonii, Vibrio diabolicus, Vibrio xuii, and V. ponticus) were obtained only at autumn, and Vibrio sinensis was only detected in winter (Supplementary Table S4). Based on the Shannon diversity index, Vibrio community at spring (H’ = 2.64 ± 0.12) was more diverse than summer (H’ = 1.90 ± 0.23) (p = 0.05). To identify specific environmental factors that may shape the community structure, RDA was conducted. RDA1 explained 62.43% of the total variation, while RDA2 explained 24.72% (Supplementary Figure S6). Only nitrate NO3– (F = 2.5, p < 0.05) was the most important factor that influenced the Vibrio diversity.
Discussion
Dongshan Bay is a semi-enclosed bay on the southeast coast, and it is an important aquaculture base for Fujian Province. The genus Vibrio is frequently associated with both wild and farmed marine animals for the ability of some species as the leading cause of foodborne outbreaks (Alam et al., 2003; Lee et al., 2008; Liu et al., 2017) and is also considered a significant problem with severe economic losses in aquaculture industry worldwide (Mancuso et al., 2015). Various studies aimed at understanding the Vibrio biogeography patterns and environmental population temporal dynamics have focused mostly on coastal and estuarine ecosystems, which suffer less human influence (Mansergh and Zehr, 2014; Westrich et al., 2016, 2018). However, current information about comprehensive spatiotemporal distributions and environmental dependencies of Vibrio communities and abundance in these aquaculture ecosystems is still limited. Overall, our results characterized that Vibrio communities in the Dongshan Bay were strongly impacted by the local seasonality, such as the fluctuations of temperature and physiochemical characters. This finding provided insights into response to environmental change, which can potentially be used to take control of blooms by interventional strategies.
Apparent seasonal variations in microbial community structure have been shown in diverse coastal ecosystems (Kirchman et al., 2005; Gilbert et al., 2012; Lau et al., 2013; Suh et al., 2015; Wang et al., 2019). The abundance and community composition of Vibrio populations in marine environments are shaped by various environmental parameters, notably temperature, salinity, and DOC (Takemura et al., 2014; Siboni et al., 2016; Kopprio et al., 2017; Zhang X.H. et al., 2018). Our results demonstrated that Vibrio diversity and abundance typically fluctuate seasonally, that the temperature had a positive correlation with the Vibrio community, and that either salinity or pH had no correlation with it.
The abundance of Vibrio spp. also showed obvious differences between aquaculture-influenced zones within the same season, especially between the FF zone and CN zone (p < 0.01), which is largely due to the difference between the amount of nutrient input in the fish farm (Fig. S2). This significant difference suggested a direct correlation between the distribution of Vibrio with the nearby aquaculture zone, which mainly farms the abalone. Many previous studies have reported the impacts of aquaculture activities on the microbial biogeography targeting the total bacterial population in the coastal environment (Pereira et al.2011; Sundberg et al.2016; Prado et al., 2020), and thus, specific lineages have been often neglected. Our results suggest that a specific phylogenetic group, that is, Vibrio, can also exhibit a clear spatial distribution pattern within the season, which is consistent with evidence by Dowle et al. (2015) and Zeng et al. (2019), who reported that total bacterial diversity changed with distance away from aquaculture zones. Besides, bacterial 16S rRNA is two to three orders of magnitude higher than the Vibrio-specific 16S rRNA, except for one sample in the fish farm at winter DY3 with only two hundred thousandths. These results are consistent with the research by Thompson et al. (2004), which found that the relative abundance of Vibrio was generally <2% of the total bacterioplankton.
A total of 167 Vibrio strains (belonging to 28 species) were identified, whose species richness was also consistent with that of other studies using culture-dependent approaches and the majority of Vibrio spp. isolates previously reported (Tall et al., 2013; Siboni et al., 2016; Wong et al., 2019). However, compared with Vibrio diversity in other temperate and subtropical coastal environments in South China Sea (Li et al., 2019; Möller et al., 2020), our results showed a differently structured Vibrio community. The diversity of Vibrio was higher in winter than in summer, which is consistent with the previous study in the water column of northern Chinese marginal seas (Liang et al., 2019). But these results are in contrast to the results of a previous study in which Vibrio was reported to be more diverse in summer than in winter (Siboni et al., 2016). The discrepancy between this previous study and our present study may be due to both that there was an actual change in the occurrence rates and that the nutrient concentrations of aquaculture from artificially affecting were different over the different regions.
Seasonality has a complex jointly impact on many hydrological gradients and inorganic and organic nutrients, which may alter the Vibrio diversity, which is also the most commonly cited reason in other reports about the Vibrio community structure change (Zhang X.H. et al., 2018; Liang et al., 2019). Furthermore, the second and third strongest environmental parameters (salinity and pH, respectively) that correlate with Vibrio diversity also do not appear to be involved in the present study. Measured salinity and pH have remained changed slightly. On the other hand, DO concentration decreased from winter to autumn, which should be associated with a subsequent decrease in Vibrio spp. In terms of inorganic and organic nutrients, NOx and PO43– were found to have correlated with the change in Vibrio spp. There was a marked decrease in NOx in the same years as the largest increases in Vibrio spp. In our study, Vibrio spp. abundance was found to increase with concentrations of NO2– and NO3–, which is consistent with observations in the Arabian Sea and Sydney Harbor estuary that spatial and temporal shifts in the abundance of the Vibrio community between seasons are primarily driven by the change of NO3– and NO2– (Asplund et al., 2011; Siboni et al., 2016). Similar relevant relationships between environmental factors and the composition of the Vibrio community were also observed with the most abundant OTU (V. fortis OTU2420), which comprised up to 43.2% of the community, and significantly correlated with DO, NO2–, and NO3–. These nutrition factors, including NOx and phosphate PO43–, were found to have correlated with the change of Vibrio spp. Contraction; a recent study suggested that they were more likely a result of the increased bacterial population, rather than a cause of nutrition triggers (Froelich et al., 2019).
Furthermore, the frequently occurring Vibrio spp. in the present study were Vibrio fortis, Vibrio alginolyticus, and Vibrio hyugaensis, which suggested that the predominance of a specific Vibrio sp. seems to be site-specific as others have reported different predominant Vibrio species in their studies, such as V. alginolyticus, Vibrio campbellii, Vibrio carchariae, Vibrio harveyi, Vibrio owensii, Vibrio parahaemolyticus, Vibrio rotiferianus, and Vibrio splendidus (Maeda et al., 2003; Eiler et al., 2006; Ortiz-Carrillo et al., 2015; Li et al., 2019; Wong et al., 2019). The four major clinically important pathogenic Vibrio species, Vibrio cholerae, V. parahaemolyticus, V. alginolyticus, and Vibrio vulnificus, have been implicated in human diseases such as gastroenteritis and septicemia and even death in immunocompromised patients (Novoslavskij et al., 2016). In the present study, the occurrence rate of V. alginolyticus was much higher than that of V. parahaemolyticus, whereas V. cholerae and V. vulnificus were not isolated, but these two lost Vibrio spp. have been extracted from oysters Crassostrea commercialis farmed at estuaries along the east coast of Australia (Siboni et al., 2016). They may have been contaminated by human fecal material, and these differences may also be explained by the halophilism of the four pathogens or their low growth rates in different nutrient conditions (Wong et al., 2019; Yan et al., 2019). Moreover, other two Vibrio species (Vibrio mimicus and Vibrio furnissii) that may have the pathogenic potential for humans were isolated from the seawater in our study (Zhang et al., 2015), and two marine animal pathogens V. harveyi and V. furnissii were also recovered by metabarcoding (Matteucci et al., 2015).
The most predominant Vibrio sp. in our study is V. fortis, which differed from Vibrio lentus in the Neuse River Estuary (Jesser and Noble, 2018) and V. campbellii, Vibrio caribbeanicus, or Vibrio atlanticus in the water column from the Chinese marginal seas (Liang et al., 2019). Recently, the effects of water temperature were examined on the microbiome of Pacific oyster Crassostrea gigas by using an experiment designed to replicate the effect of a marine heat wave event, yet this study found that heat stress increased the abundance of V. fortis by 10-fold (Green et al., 2019), indicating that the growth of V. fortis has been promoted by warmer temperatures, and they may represent warm-water species.
In addition, another paradoxical and impenetrable phenomenon is that the disappearance of the summer predominant Vibrio species may be associated with the high diversity of Vibrio that was observed in winter. One reason is that the food supply for aquaculture animals has introduced more nutrients that favor primary productivity and subsequently the growth of heterotrophic bacteria, including Vibrio spp. Most studies on Vibrio diversity have an agreement that many Vibrio spp. enter viable but non-culturable growth states (VBNCs) when exposed to adverse growth conditions will underestimate Vibrio diversity, as we know that the water temperatures that increased the effect of VBNC on our Vibrio spp. counts were presumed to be negligible (Oliver, 2010; Vezzulli et al., 2012, 2013; Takemura et al., 2014).
After identification of OTUs in SILVA16S rRNA database, taxonomy was also reassigned against the EzBioCloud database, which will give more information about the latest taxonomy of microbial isolates and more complete taxonomic hierarchy from phylum to species for the isolates (Quast et al., 2012; Yoon et al.2017). Several OTUs annotated as Vibrio spp. (OTU967, OTU1721, OTU2461, OTU1824, OTU1583, OTU2611, OTU1776, OTU2096, OTU2124, OTU499, and OTU489) in SILVA were assigned to Vibrio maritimus, Vibrio zhanjiangensis, V. maritimus, Vibrio aestivus, Vibrio echinoideorum, Catenococcus thiocycli, Vibrio cortegadensis, V. rotiferianus, Vibrio hepatarius, V. caribbeanicus, V. furnissii, and also three OTUs (OTU607, OTU1812, and OTU3517) cannot be affiliated to species level. It was reported that universal 16S rRNA primers may have some limitations; the most important fact is that it does not contain highly variable domains that are probably required to analyze relationships of closely related organisms, which indicated that it was necessary for Vibrio to develop a high species diversity, such as Vibrio-specific 16S rRNA and hsp60 sequencing retrieved more than did the present study (Siboni et al., 2016; Jesser and Noble, 2018; Liang et al., 2019).
Conclusion
This study investigated the Vibrio communities within aquaculture zone in relation to the established environment factors based on 1-year survey. This observation demonstrated the importance of seasonal pattern in determining the Vibrio abundance and community composition on these three different zones, temporal-spatial assemblages on these three different zones, where significant abundances of Vibrio already occur, are potentially at risk of pathogenic Vibrio outbreaks, particularly during warm summer months. Both the abundance and community composition varied between different sea areas, with the Vibrio assemblages displaying a clear distance-decay pattern. In addition, Vibrio fortis was dominant in these aquaculture areas by universal bacteria 16S rRNA gene and cultivation methods. Our present study is one of the few studies to examine the occurrence of Vibrio in the marine aquaculture zone of South China, and the results indicate that Vibrio are widely distributed in aquaculture environment and that a further risk assessment is needed to be conducted.
Data Availability Statement
The raw sequencing reads have been submitted to the Sequence Read Archive (SRA) database with accession number SRR11929868–SRR11929907.
Author Contributions
WX is responsible for article writing and data analysis. LG, XM, and ZL are responsible for data analysis. SY and YG are responsible for sample collection. LX is responsible for experiment. CS is responsible for sample collection. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The work was financially supported by the National Key R&D Program of China under Contract (No. 2017YFC1404500), the Scientific Research Foundation of Third Institute of Oceanography, MNR (No. 2019022), China-ASEAN Maritime Cooperation Fund Project “Monitoring and conservation of the coastal ecosystem in the South China Sea”, National Natural Science Foundation of China (Nos. 41776170 and 91951102), Natural Science Foundation of Fujian Province (No. 26185006), Bilateral Cooperation of Maritime Affairs (No. HC01-200302), and Science and Technology Innovation Committee of Shenzhen (Nos. JCYJ20170818091727570 and JCYJ20190808152403587).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.575287/full#supplementary-material
Clustering analysis of the normalized environmental parameters using the average cluster method.
Vibrio abundance determined by qPCR in the three different sample areas (fish farm, water channel of farm zone and control zone). The asterisks denote significant differences between seasons. *P ¡ 0.05, **PP ¡ 0.01.
Venn diagram showing the OTUs that obtained and overlap among total samples across the four seasons.
Vibrio community structure determined by dominant OTUs counted in the 40 water samples.
Venn diagram showing the OTUs that obtained and overlap among total samples within three different zones.
RDA biplots for four seasons based on Vibrio isolates data and environmental variables.
Environmental parameters of all sampling sites across the four seasons.
Observed reads and diversity estimates of Vibrio spp. based on 97% OTU clusters.
Classification of obtained 22 Vibrio OTUs based on three different database.
Classification of obtained 23 Vibrio species based on 16S rRNA sequences.
References
- Alam M. J., Miyoshi S., Shinoda S. (2003). Studies on pathogenic Vibrio parahaemolyticus during a warm weather season in the Seto Inland Sea. Japan. Environ. Microbiol. 5 706–710. 10.1046/j.1462-2920.2003.00458.x [DOI] [PubMed] [Google Scholar]
- Amin A., Feng G., Al-saari N., Meirelles P., Yamazaki Y., Mino S., et al. (2016). The first temporal and spatial assessment of Vibrio diversity of the surrounding seawater of coral reefs in ishigaki. Japan. Front. Microbiol. 7:1185. 10.3389/fmicb.2016.01185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asplund M. E., Rehnstam-Holm A. S., Atnur V., Raghunath P., Saravanan V., Harnstrom K., et al. (2011). Water column dynamics of Vibrio in relation to phytoplankton community composition and environmental conditions in a tropical coastal area. Environ. Microbiol. 13 2738–2751. 10.1111/j.1462-2920.2011.02545.x [DOI] [PubMed] [Google Scholar]
- Baffone W., Tarsi R., Pane L., Campana R., Repetto B., Mariottini G. L., et al. (2006). Detection of free-living and plankton-bound vibrios in coastal waters of the Adriatic Sea (Italy) and study of their pathogenicity-associated properties. Environ. Microbiol. 8 1299–1305. 10.1111/j.1462-2920.2006.01011.x [DOI] [PubMed] [Google Scholar]
- Dowle E., Pochon X., Keeley N., Wood S. A. (2015). Assessing the effects of salmon farming seabed enrichment using bacterial community diversity and high-throughput sequencing. FEMS Microbiol. Ecol. 91:fiv089. 10.1093/femsec/fiv089 [DOI] [PubMed] [Google Scholar]
- Eiler A., Johansson M., Bertilsson S. (2006). Environmental influences on Vibrio populations in northern temperate and boreal coastal waters (Baltic and Skagerrak Seas). Appl. Environ. Microbiol. 72 6004–6011. 10.1128/AEM.00917-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonte E. S., Amado A. M., Meirelles-Pereira F., Esteves F. A., Rosado A. S., Farjalla V. F. (2013). The combination of different carbon sources enhances bacterial growth efficiency in aquatic ecosystems. Microbiol. Ecol. 66 871–878. 10.1007/s00248-013-0277-1 [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations [FAO], and The State of World Fisheries and Aquaculture [WFA] (2016). Contributing to Food Security and Nutrition for All, Report (Rome, 2016). Available from http://www.fao.org/fishery/sofia/en. [Google Scholar]
- Froelich B., Gonzalez R., Blackwood D., Lauer K., Noble R. (2019). Decadal monitoring reveals an increase in Vibrio spp. concentrations in the Neuse River Estuary. North Carolina, USA. PLoS One 14:e0215254. 10.1371/journal.pone.0215254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J. A., Steele J. A., Caporaso J. G., Steinbruck L., Reeder J., Temperton B., et al. (2012). Defining seasonal marine microbial community dynamics. ISME J. 6 298–308. 10.1038/ismej.2011.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T. J., Siboni N., King W. L., Labbate M., Seymour J. R., Raftos D. (2019). Simulated marine heat wave alters abundance and structure of Vibrio populations associated with the Pacific Oyster resulting in a mass mortality event. Microbiol. Ecol. 77 736–747. 10.1007/s00248-018-1242-9 [DOI] [PubMed] [Google Scholar]
- Guo M., Wu F., Hao G., Qi Q., Li R., Li N. (2017). Bacillus subtilis improves immunity and disease resistance in rabbits. Front. Immunol. 8:354. 10.3389/fimmu.2017.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall E. K., Neuhauser C., Cotner J. B. (2008). Toward a mechanistic understanding of how natural bacterial communities respond to changes in temperature in aquatic ecosystems. ISME J. 2 471–481. 10.1038/ismej.2008.9 [DOI] [PubMed] [Google Scholar]
- Hayashi H., Sakamoto M., Benno Y. (2002). Phylogenetic analysis of the human gut microbiota using 16s rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46 535–548. 10.1111/j.1348-0421.2002.tb02731.x [DOI] [PubMed] [Google Scholar]
- Hsieh J. L., Fries J. S., Noble R. T. (2008). Dynamics and predictive modelling of Vibrio spp. in the Neuse river estuary, North Carolina, USA. Environ, Microbiol. 10 57–64. 10.1111/j.1462-2920.2007.01429.x [DOI] [PubMed] [Google Scholar]
- Jesser K. J., Noble R. T. (2018). Characterizing the ecology of Vibrio in the Neuse River Estuary, North Carolina using heat shock protein 60 (hsp60) next-generation amplicon sequencing. Appl. Environ. Microbiol. 84:e00333-18. 10.1128/AEM.00333-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julie D., Solen L., Antoine V., Jaufrey C., Annick D., Dominique H. H., et al. (2010). Ecology of pathogenic and non-pathogenic Vibrio parahaemolyticus on the French Atlantic coast. Effects of temperature, salinity, turbidity and chlorophyll a. Environ. Microbiol. 12 929–937. 10.1111/j.1462-2920.2009.02136.x [DOI] [PubMed] [Google Scholar]
- Kirchman D. L., Dittel A. I., Malmstrom R. R., Cottrell M. T. (2005). Biogeography of major bacterial groups in the Delaware Estuary. Limnol. Oceanogr. 50 1697–1706. 10.4319/lo.2005.50.5.1697 [DOI] [Google Scholar]
- Kopprio G. A., Streitenberger M. E., Okuno K., Baldini M., Biancalana F., Fricke A., et al. (2017). Biogeochemical and hydrological drivers of the dynamics of Vibrio species in two Patagonian estuaries. Sci. Total Environ. 579 646–656. 10.1016/j.scitotenv.2016.11.045 [DOI] [PubMed] [Google Scholar]
- Lau S. C., Zhang R., Brodie E. L., Piceno Y. M., Andersen G., Liu W. T. (2013). Biogeography of bacterioplankton in the tropical seawaters of Singapore. FEMS Microbiol. Ecol. 84 259–269. 10.1111/1574-6941.12057 [DOI] [PubMed] [Google Scholar]
- Lee J. K., Jung D. W., Eom S. Y., Oh S. W., Kim Y., Kwak H. S., et al. (2008). Occurrence of Vibrio parahaemolyticus in oysters from Korean retail outlets. Food Control. 19 990–994. 10.1016/j.foodcont.2007.10.006 [DOI] [Google Scholar]
- Lee L. H., Raghunath P. (2018). Vibrionaceae diversity, multidrug resistance and management. Front. Microbiol. 9:563 10.3389/978-2-88945-476-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Liu J., Zhou S., Fu L., Zhang X. H. (2019). Vertical variation in Vibrio community composition in sansha yongle blue hole and its ability to degrade macromolecules. Mar. Life Sci. Technol. 2 60–72. 10.1007/s42995-019-00003-4 [DOI] [Google Scholar]
- Liang J., Liu J., Wang X., Lin H., Liu J., Zhou S., et al. (2019). Spatiotemporal dynamics of free-living and particle-associated Vibrio communities in the northern Chinese marginal seas. Appl. Environ. Microbiol. 85 e217–e219. 10.1128/AEM.00217-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bai L., Li W., Han H., Fu P., Ma X., et al. (2017). Trends of foodborne diseases in China: lessons from laboratory-based surveillance since 2011. Front. Med. 12:48–57. 10.1007/s11684-017-0608-6 [DOI] [PubMed] [Google Scholar]
- Lopez-Hernandez K. M., Pardio-Sedas V. T., Lizarraga-Partida L., Williams J., de J., Martinez-Herrera D., et al. (2015). Environmental parameters influence on the dynamics of total and pathogenic Vibrio parahaemolyticus densities in Crassostrea virginica harvested from Mexico’s Gulf coast. Mar. Pollut. Bull. 91 317–329. 10.1016/j.marpolbul.2014.11.015 [DOI] [PubMed] [Google Scholar]
- Maeda T., Matsuo Y., Furushita M., Shiba T. (2003). Seasonal dynamics in a coastal Vibrio community examined by a rapid clustering method based on 16S rDNA. Fisheries Sci. 69 385–394. 10.1046/j.1444-2906.2003.00633.x [DOI] [Google Scholar]
- Mancuso M., Genovese M., Guerrera M. C., Casella G., Genovese L., Piccolo G., et al. (2015). First episode of vibriosis in wild specimens of Pagellus bogaraveo (Brünnich, 1768) in the Mediterranean Sea. Cah. Biol. Mar. 56 355–361. 10.21411/CBM.A.B1400584 [DOI] [Google Scholar]
- Mansergh S., Zehr J. P. (2014). Vibrio diversity and dynamics in the monterey bay upwelling region. Front. Microbiol. 5:48. 10.3389/fmicb.2014.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteucci G., Schippa S., Di L. G., Migliore L., Thaller M. C. (2015). Species diversity, spatial distribution, and virulence associated genes of culturable vibrios in a brackish coastal Mediterranean environment. Ann. Microbiol. 65 2311–2321. 10.1007/s13213-015-1073-6 [DOI] [Google Scholar]
- Möller L., Kreikemeyer B., Luo Z.-H., Jost G., Labrenz M. (2020). Impact of coastal aquaculture operation systems in Hainan island (China) on the relative abundance and community structure of Vibrio in adjacent coastal systems. Estuar. Coast. Shelf Sci. 233:106542 10.1016/j.ecss.2019.106542 [DOI] [Google Scholar]
- Nigro O. D., Hou A., Vithanage G., Fujioka R. S., Steward G. F. (2011). Temporal and spatial variability in culturable pathogenic Vibrio spp. in Lake Pontchartrain, Louisiana, following hurricanes Katrina and Rita. Appl. Environ. Microb. 77 5384–5393. 10.1128/AEM.02509-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoslavskij A., Terentjeva M., Eizenberga I., Valciòa O., Bartkeviès V., Bçrziòð A. (2016). Major foodborne pathogens in fish and fish products: a review. Ann. Microbiol. 66 1–15. 10.1007/s13213-015-1102-5 [DOI] [Google Scholar]
- Oberbeckmann S., Fuchs B. M., Meiners M., Wichels A., Wiltshire K. H., Gerdts G. (2012). Seasonal dynamics and modeling of a Vibrio community in coastal waters of the North Sea. Microb. Ecol. 63 543–551. 10.1007/s00248-011-9990-9 [DOI] [PubMed] [Google Scholar]
- Oliver J. D. (2010). Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 34 415–425. 10.1111/j.1574-6976.2009.00200.x [DOI] [PubMed] [Google Scholar]
- Ortiz-Carrillo I., Estrella-Gómez N. E., Zamudio-Maya M., Rojas-Herrera R. (2015). Diversity of Vibrio in karstic coastal marshes in the Yucatan Peninsula. PLoS One 10:e0134953. 10.1371/journal.pone.0134953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira C., Salvador S., Arrojado C., Silva Y., Santos A. L., Cunha Â, et al. (2011). Evaluating seasonal dynamics of bacterial communities in marine fish aquaculture: a preliminary study before applying phage therapy. J. Environ. Monitor. 13 1053–1058. 10.1039/c0em00434k [DOI] [PubMed] [Google Scholar]
- Prado S., Barja J. L., Luzardo A., Dubert J., Blanco J. (2020). Encapsulation of live marine bacteria for use in aquaculture facilities and process evaluation using response surface methodology. Appl. Microbiol. Biot. 104 1993–2006. 10.1007/s00253-019-10332-0 [DOI] [PubMed] [Google Scholar]
- Pruzzo C., Huq A., Colwell R. R., Donelli G. (2005). “Pathogenic Vibrio species in the marine and estuarine environment,” in Oceans & Health Pathogens in the Marine Environment, eds Belkin S., Colwell R. R. (New York, NY: Springer; ), 217–252. 10.1007/0-387-23709-7_9 [DOI] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web–based tools. Nucleic Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siboni N., Balaraju V., Carney R., Labbate M., Seymour J. R. (2016). Spatiotemporal dynamics of Vibrio spp. within the sydney harbour estuary. Front. Microbiol. 7:460. 10.3389/fmicb.2016.00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh S. S., Park M., Hwang J., Kil E. J., Jung S., Lee S., et al. (2015). Seasonal dynamics of marine microbial community in the South Sea of Korea. PLoS One 10:e0131633. 10.1371/journal.pone.0131633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg L. R., Ketola T., Laanto E., Kinnula H., Bamford J. K., Penttinen R., et al. (2016). Intensive aquaculture selects for increased virulence and interference competition in bacteria. Proc. R. Soc. B 283:20153069. 10.1098/rspb.2015.3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura A. F., Chien D. M., Polz M. F. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5:38. 10.3389/fmicb.2014.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tall A., Hervio-Heath D., Teillon A., Boisset-Helbert C., Delesmont R., Bodilis J., et al. (2013). Diversity of Vibrio spp. isolated at ambient environmental temperature in the eastern English Channel as determined by pyrh sequencing. J. Appl. Microbiol. 114 1713–1724. 10.1111/jam.12181 [DOI] [PubMed] [Google Scholar]
- Thompson J. R., Polz M. F. (2006). “Dynamics of Vibrio populations and their role in environmental nutrient cycling,” in The Biology of Vibrios, eds Thompson F., Austin B., Swings J. (Washington, DC: American Society of Microbiology; ), 190–203. 10.1128/9781555815714 [DOI] [Google Scholar]
- Thompson J. R., Randa M. A., Marcelino L. A., Tomita-Mitchell A., Lim E., Polz M. F. (2004). Diversity and dynamics of a north atlantic coastal Vibrio community. Appl. Environ. Microbiol. 70 4103–4110. 10.1128/AEM.70.7.4103-4110.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L., Brettar I., Pezzati E., Reid P. C., Colwell R. R., Hofle M. G., et al. (2012). Long-term effects of ocean warming on the prokaryotic community: evidence from the vibrios. ISME J. 6 21–30. 10.1038/ismej.2011.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L., Colwell R. R., Pruzzo C. (2013). Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb. Ecol. 65 817–825. 10.1007/s00248-012-0163-2 [DOI] [PubMed] [Google Scholar]
- Vezzulli L., Grande C., Reid P. C., Helaouet P., Edwards M., Hofle M. G., et al. (2016). Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc. Natl. Acad. Sci. U.S.A. 113 E5062–E5071. 10.1073/pnas.1609157113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu Y., Wang J., Luo T., Zhang R., Sun J., et al. (2019). Seasonal dynamics of bacterial communities in the surface seawater around subtropical Xiamen Island, China, as determined by 16S rRNA gene profiling. Mar. Pollut. Bull. 142 135–144. 10.1016/j.marpolbul.2019.03.035 [DOI] [PubMed] [Google Scholar]
- Ward C. S., Yung C. M., Davis K. M., Blinebry S. K., Williams T. C., Johnson Z. I., et al. (2017). Annual community patterns are driven by seasonal switching between closely related marine bacteria. ISME J. 11 1412–1422. 10.1038/ismej.2017.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich J. R., Ebling A. M., Landing W. M., Joyner J. L., Kemp K. M., Griffin D. W., et al. (2016). Saharan dust nutrients promote Vibrio bloom formation in marine surface waters. Proc. Natl. Acad. Sci. U.S.A. 113 5964–5969. 10.1073/pnas.1518080113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrich J. R., Griffin D. W., Westphal D. L., Lipp E. K. (2018). Vibrio population dynamics in mid-atlantic surface waters during saharan dust events. Front. Mar. Sci. 5:12 10.3389/fmars.2018.00012 [DOI] [Google Scholar]
- Wong Y. Y., Lee C. W., Bong C. W., Lim J. H., Narayanan K., Sim E. U. H. (2019). Environmental control of Vibrio spp. abundance and community structure in tropical waters. FEMS Microbiol. Ecol. 95:fiz176. 10.1093/femsec/fiz176 [DOI] [PubMed] [Google Scholar]
- Yan L., Pei X., Zhang X., Guan W., Chui H., Jia H., et al. (2019). Occurrence of four pathogenic Vibrios in Chinese freshwater fish farms in 2016. Food Control 95 85–89. 10.1016/j.foodcont.2018.07.043 [DOI] [Google Scholar]
- Yoon S. H., Ha S. M., Kwon S., Lim J., Kim Y., Seo H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67 1613–1617. 10.1099/ijsem.0.001755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J., Lin Y., Zhao D., Huang R., Xu H., Jiao C. (2019). Seasonality overwhelms aquacultural activity in determining the composition and assembly of the bacterial community in Lake Taihu, china. Sci. Total Environ. 683 427–435. 10.1016/j.scitotenv.2019.05.256 [DOI] [PubMed] [Google Scholar]
- Zhang W., Mo Y., Yang J., Zhou J., Lin Y., Isabwe A., et al. (2018). Genetic diversity pattern of microeukaryotic communities and its relationship with the environment based on PCR-DGGE and T-RFLP techniques in Dongshan Bay, southeast China. Cont Shelf Res. 164 1–9. 10.1016/j.csr.2018.05.006 [DOI] [Google Scholar]
- Zhang X. H., Lin H., Wang X., Brian A. (2018). Significance of Vibrio species in the marine organic carbon cycle—a review. Sci. China Earth Sci. 61 1357–1368. 10.1007/s11430-017-9229-x [DOI] [Google Scholar]
- Zhang X. F., Li K., Wu S., Shuai J. B., Fang W. H. (2015). Peptide nucleic acid fluorescence in-situ hybridization for identification of Vibrio spp. in aquatic products and environments. Int. J. Food Microbiol. 206 39–44. 10.1016/j.ijfoodmicro.2015.04.017 [DOI] [PubMed] [Google Scholar]
- Zhu C., Zhang J., Nawaz M. Z., Mahboob S., Al-Ghanim K. A., Khan I. A., et al. (2019). Seasonal succession and spatial distribution of bacterial community structure in a eutrophic freshwater Lake, Lake Taihu. Sci. Total Environ. 669 29–40. 10.1016/j.scitotenv.2019.03.087 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clustering analysis of the normalized environmental parameters using the average cluster method.
Vibrio abundance determined by qPCR in the three different sample areas (fish farm, water channel of farm zone and control zone). The asterisks denote significant differences between seasons. *P ¡ 0.05, **PP ¡ 0.01.
Venn diagram showing the OTUs that obtained and overlap among total samples across the four seasons.
Vibrio community structure determined by dominant OTUs counted in the 40 water samples.
Venn diagram showing the OTUs that obtained and overlap among total samples within three different zones.
RDA biplots for four seasons based on Vibrio isolates data and environmental variables.
Environmental parameters of all sampling sites across the four seasons.
Observed reads and diversity estimates of Vibrio spp. based on 97% OTU clusters.
Classification of obtained 22 Vibrio OTUs based on three different database.
Classification of obtained 23 Vibrio species based on 16S rRNA sequences.
Data Availability Statement
The raw sequencing reads have been submitted to the Sequence Read Archive (SRA) database with accession number SRR11929868–SRR11929907.


