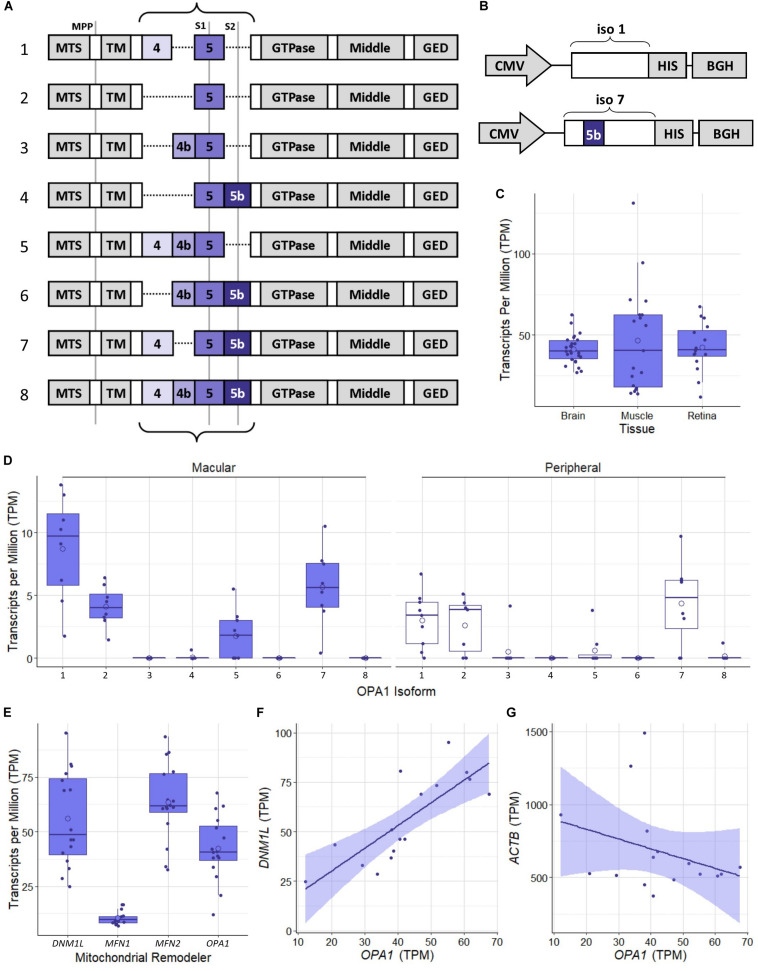
FIGURE 1.
Schematic representation of OPA1 RNA isoforms and RNAseq analysis of healthy human tissue. (A) The 8 primary protein coding mRNA variants of OPA1 in humans. The curly braces indicate the area that undergoes alternate splicing of exons 4, 4b, and 5b. Gray lines indicate protease cleavage sites, MPP = matrix processing peptidases, S1 = OMA1 and S2 = YME1L. Gray blocks outside of the curly braces indicate functional domains; MTS, Mitochondrial Targeting Sequence; TM, Transmembrane domain; GED, GTPase effector domain. (B) Schematics of optimized OPA1 isoform 1 or 7 expression cassettes. CMV, Cytomegalovirus promoter; HIS, 6x HIS tag; BGH, Bovine Growth Hormone polyadenylation signal. Note that only exon 5b differs between them. (C) Comparison of OPA1 transcript levels in Transcripts Per Million (TPM) in healthy dorsolateral prefrontal cortex brain tissue (n = 26 samples), skeletal muscle tissue (n = 19) and retinal tissue (n = 16). The open circle on each boxplot represents the mean. (D) Expression levels of the 8 protein coding isoforms of OPA1 as identified in the ensemble database (ENSG00000198836) measured in TPM. n = 8 macular samples and n = 8 peripheral samples. (E) RNA transcript expression levels in TPM of 4 significant mitochondrial remodeling proteins Drp1 (DNM1L), Mitofusin 1 and 2 (MFN1 and 2) and OPA1. n = 16 (macular and peripheral) samples. (F,G) Are representative scatter plots of the correlation between mitochondrial remodeler RNA transcripts, with ACTB as a control. OPA1 transcript expression is compared to either DNM1L or ACTB transcript levels, measured in TPM. DNM1L and OPA1 transcript levels show a significant positive correlation (r = 0.79, Pearson’s Correlation Coefficient, n = 16, p < 0.05), whereas ACTB and OPA1 transcript levels show no significant correlation (r = –0.32, Pearson’s Correlation Coefficient, n = 16, p > 0.05). Shaded region represents 95% confidence interval, BH correction for multiple testing.