Figure 7.
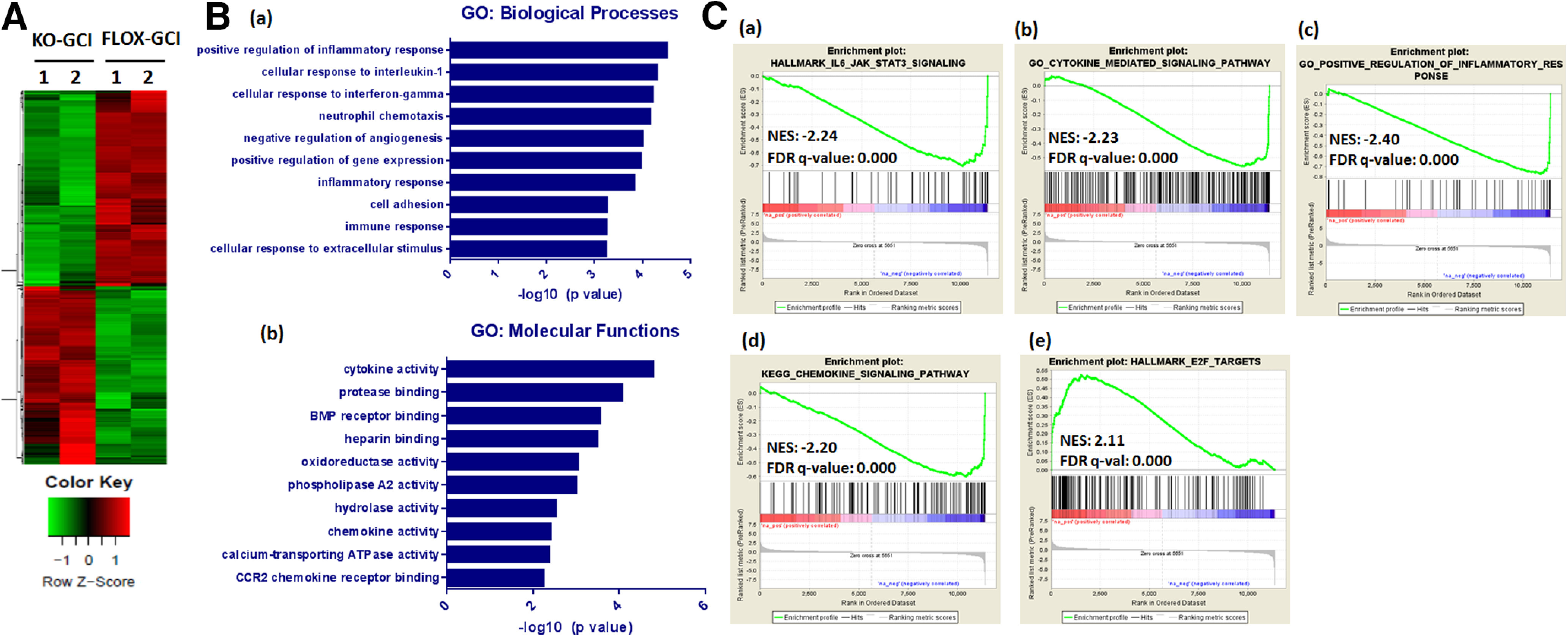
RNA-seq analysis of hippocampal transcriptome alterations in ovx-female GFAP-ARO-KO mice after GCI. A, Heat map of DEGs between FLOX-GCI and KO-GCI groups at 24 h after GCI is shown (n = 2, >2-fold, p < 0.05). B, GO analysis of DEGs in terms of biological processes [B(a)] or molecular functions [B(b)] was examined using DAVID software and the top ten significantly altered pathways are shown. C, GSEA revealed that the IL-6/JAK/STAT3 signaling [C(a)], cytokine mediated signaling pathway [C(b)], positive regulation of inflammatory response [C(c)], and chemokine-signaling pathway gene signatures [C(d)] showed negative correlation with genes altered in KO-GCI-R24h; E2F-targets [C(e)] showed positive correlation with genes altered in KO-GCI-R24h. NES, normalized enrichment score.
