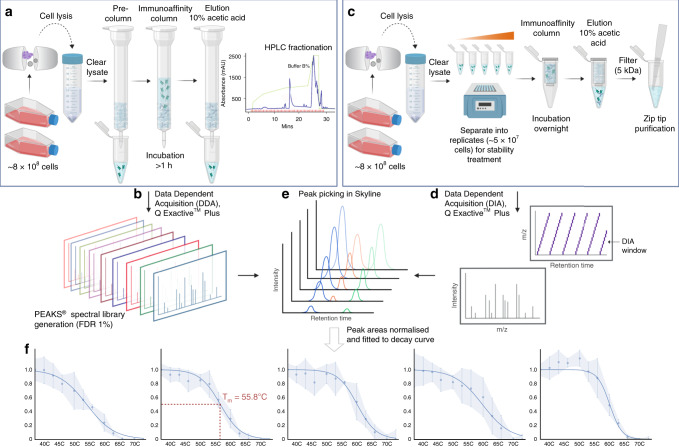
Fig. 1. Workflow for stability profiling of HLA-associated peptides.
a Initially, immunoprecipitation on C1R cells expressing the HLA allele of interest was carried out by culturing cells, lysing them, clearing the lysate, and isolating peptides according to established workflows9. b Liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis of pHLA eluates was performed in DDA mode to create HLA allele-specific spectral libraries. c Small-scale immunoprecipitation was carried out by clearing lysates and separating these into replicates consisting of 5 × 107 cells, after which aliquots were incubated in triplicate at temperatures ranging from 37 to 73 °C with a temperature step-size of 3-4 °C. d Subsequently, the remaining thermostable HLA-bound peptides were eluted, filtered, and analyzed using a DIA strategy to enable peptide quantification at different temperature points. e Spectral library matching and filtering were performed in Skyline. f Peptide peak areas for triplicate samples were normalized and fitted to sigmoidal decay curves to determine the temperature at which half of the complex was unfolded, termed the thermal melting temperature (Tm). Thermal treatment of C1R cell lysates was carried out at 12 different temperature points ranging from 37 to 73 °C with n=3 biological replicates at each temperature point. Data are presented as median values ± SD. FDR: False Discovery Rate. Parts of the figure were generated using BioRender.com.