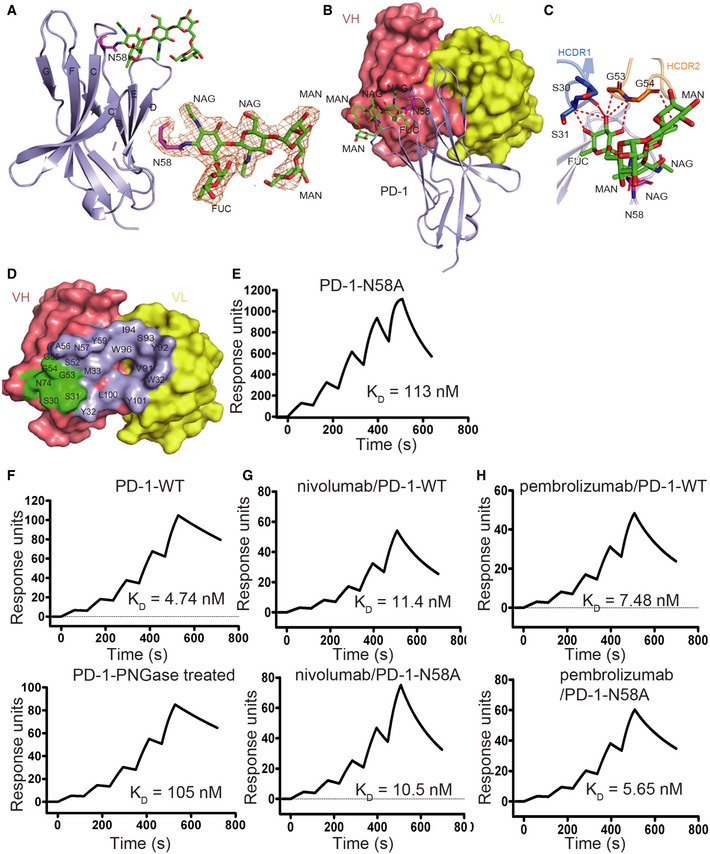
-
A
The overall structure of N‐glycan on N58 of PD‐1 is presented with N‐glycans on N58 of PD‐1 highlighted in sticks in green. The 2 Fo‐Fc electron density map of the N58 N‐glycans contoured at 1.0 sigma is represented in orange on the right. The β strands were labeled according to canonical IgV‐strand designations.
-
B
Location of N58 glycosylation in the interaction with camrelizumab‐scFv. VL of camrelizumab‐scFv is colored in yellow and VH in deep salmon.
-
C
Detailed interactions of N‐glycans on N58 of PD‐1 with camrelizumab‐scFv. The observed glycans on N58 of PD‐1 are highlighted in sticks and colored in green. Residues with hydrogen bond and van der Waals force interaction are shown as sticks and labeled while Hydrogen bond and van der Waals force interaction are shown as dashed red lines.
-
D
Binding paratope on VH and VL of camrelizumab by PD‐1 is presented with residues in contact with glycan of N58 highlighted in green.
-
E
SPR analysis of the binding of camrelizumab with N58A mutated PD‐1 expressed in 293T cells.
-
F
The SPR analysis of the binding of camrelizumab with WT (upper) or PNGase F treated PD‐1 proteins (lower) expressed in 293T cells.
-
G, H
The SPR analysis of the binding of nivolumab (G) with WT (upper) or N58A mutated (lower) PD‐1 expressed in 293T cells, and the binding of pembrolizumab (H) with WT (upper) or N58A‐mutated (lower) PD‐1 proteins.