Figure 2. Effects of Dronc deficiency in intestinal stem cells in experimental conditions without basal cellular turnover.
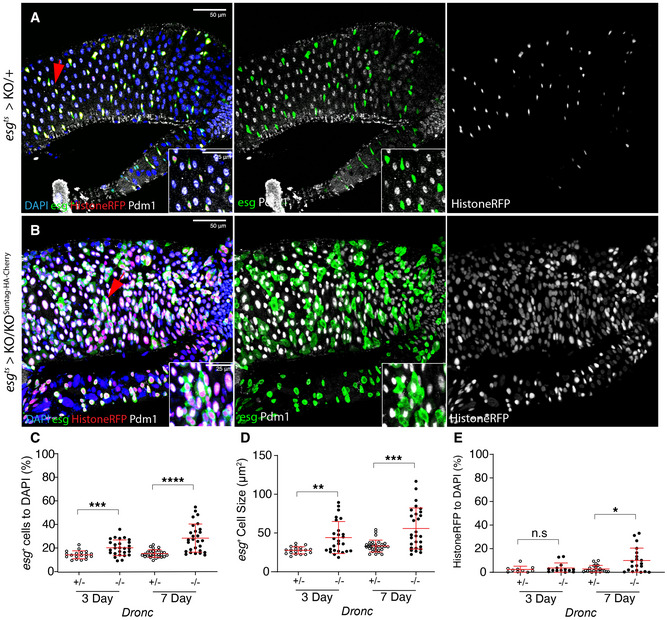
- ReDDM activation in Dronc heterozygous (+/−) intestine 7 days after temperature shift at 29°C; esg expression (green) labels the intestinal progenitor cells, Histone‐RFP (red) is a semi‐permanent marker retained in differentiated cells and Pdm‐1 (grey) labels differentiated ECs. The red arrows indicate the enlarged areas depicted in the insets in the entire figure. DAPI labels the DNA In the entire Figure (blue). All of the experiments in the entire figure have been made under experimental conditions that protect the epithelial integrity (intestine reared in Oxford Medium and flies transferred every 2 days into vials with fresh food). Genotype: w 1118; esg‐Gal4 UAS‐CD8‐GFP/+; TubG80ts UAS‐Histone‐RFP Dronc KO (recombinant chromosome made for this study)/+.
- ReDDM activation in a Drosophila intestine in which all of the progenitor cells are fully mutant for Dronc (see full genotype in Materials and Methods); note the increased cell size of esg‐expressing cells (GFP+, green), the co‐expression of Histone‐RFP (red) and the EC marker Pdm‐1 (grey), and the co‐expression of Pdm‐1 and GFP in enlarged cells (compare panels and detailed insets from A with B). The image is a representative example of a Drosophila adult gut 7 days after temperature shift at 29°C. Genotype: w 1118; esg‐Gal4 UAS‐CD8‐GFP/+; TubG80ts UAS‐Histone‐RFP Dronc KO/UAS‐Flippase (BL8209) FRT Dronc‐GFP‐APEX FRT suntag‐HA‐Cherry.
- Relative number of esg‐expressing cells normalised to DAPI; notice that the relative percentage of esg‐labelled cells is significantly higher in Dronc fully mutant conditions (−/−) at 3 days (***P = 0.0007) and 7d (****P < 0.0001) post‐temperature shift at 29°C (Quantifications were made using N ≥ 2 biological replicates; unpaired two‐tailed t‐test, +/− n = 32, −/− n = 25). Error bars represent standard deviation of the mean. Genotypes: +/−: w 1118; esg‐Gal4 UAS‐CD8‐GFP/+; TubG80ts UAS‐Histone‐RFP Dronc KO/+; −/−: w 1118; esg‐Gal4 UAS‐CD8‐GFP/+; TubG80ts UAS‐Histone‐RFP Dronc KO/UAS‐Flippase (BL8209) FRT Dronc‐GFP‐APEX FRT suntag‐HA‐Cherry.
- Average cell size of esg‐expressing cells (μm2); note the increased cell size of Dronc fully mutant progenitor cells (−/−) at 3d (**P = 0.0014) and 7d (***P = 0.0007) post‐temperature shift at 29°C (Quantifications were made using N ≥ 2 biological replicates; unpaired two‐tailed parametric t‐test, 3d +/− n = 19, 3d −/− n = 28, 7d +/− n = 29, 7d −/− n = 28). Error bars represent standard deviation of the mean. Genotypes: +/−: w 1118; esg‐Gal4 UAS‐CD8‐GFP/+; TubG80ts UAS‐Histone‐RFP Dronc KO/+; −/−: w 1118; esg‐Gal4 UAS‐CD8‐GFP/ +; TubG80ts UAS‐Histone‐RFP Dronc KO/ UAS‐Flippase (BL8209) FRT Dronc‐GFP‐APEX FRT suntag‐HA‐Cherry.
- Relative number of esg‐negative cells expressing Histone‐RFP normalised to DAPI; notice that the number of Histone‐RFP cells without esg expression is significantly higher in Dronc fully mutant (−/−) conditions at 7d (*P = 0.0046) (Quantifications were made using N biological replicates ≥2; Mann–Whitney test, +/− n = 24, −/− n = 20. Error bars represent standard deviation of the mean. Genotypes: +/−: w 1118; esg‐Gal4 UAS‐CD8‐GFP/+; TubG80ts UAS‐Histone‐RFP Dronc KO/+; −/−: w 1118; esg‐Gal4 UAS‐CD8‐GFP/+; TubG80ts UAS‐Histone‐RFP Dronc KO/UAS‐Flippase (BL8209) FRT Dronc‐GFP‐APEX FRT suntag‐HA‐Cherry.
Data information: (A, B) The red arrows indicate the enlarged areas depicted in the insets in the entire figure.
