Abstract
This year’s Nobel prize for the CRISPR/Cas system is an illustrative example of how scientific breakthroughs rests on preceding work: the discovery of guide RNAs in the 1990s.
Subject Categories: S&S: History & Philosophy of Science, RNA Biology
No one studying molecular biology can escape the impact that the CRISPR/Cas9 system has had on the field. In its natural environment, it functions as an adaptive bacterial immune system that can remember past viral infections. However, it is mainly known and popular as a highly versatile and precise tool for gene editing, the development of which was awarded this year with the Nobel Price for chemistry for Emmanuelle Charpentier and Jennifer Doudna (Jinek, Chylinski et al, 2012). At the heart of the system is Cas9, a programmable and sequence‐specific endonuclease that introduces double‐strand breaks into DNA, which can be exploited to modify the DNA sequence in many different ways. The Cas9 cleavage site is determined by a small RNA molecule, termed guide RNA (gRNA), that base pairs with the target DNA and at the same time binds to Cas9 to initiate the site‐specific double‐strand break (Fig. 1). What is not generally known is that the discovery of the first gRNAs and the concept that a small RNA directs a protein activity to a defined nucleic acid sequence by base pairing predates the discovery of the CRISPR/Cas9 system by more than two decades.
Figure 1. Schematic comparisons of four groups of “gRNAs” in chronological order of their discovery.
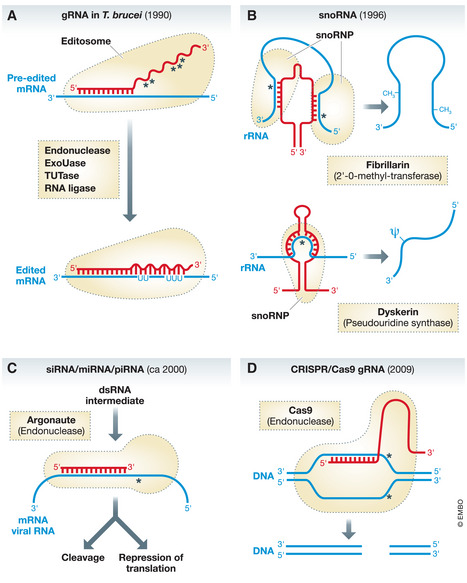
(A) gRNAs in trypanosomes mediate mitochondrial RNA editing. They determine the sites where uridines are inserted and/or deleted from the pre‐edited RNA. Editing requires sequential reactions of an endoribonuclease (endonuclease), terminal uridyltransferase (Tutase), U‐specific exoribonuclease (ExoUase), and an RNA ligase. Wavy red line indicates the gRNA sequence that mediates RNA editing. (B) Two different types of snoRNA mediate rRNA 2’‐O‐methylation and pseudouridylation, respectively. (C) siRNA and miRNA are processed by distinct pathways resulting in a double‐stranded RNA intermediate, the sense strand of which gets incorporated into a protein complex that includes a member of the argonaute protein family. siRNA, miRNA, and piRNA mediate gene expression by degradation of their target RNA or by repression or activation of translation and transcription. (D) The CRISPR/Cas9 system mediates a double‐strand DNA break whose position is determined by the gRNA. The system is presently mainly known for simplifying many forms of genetic engineering in a wide variety of systems. The picture here shows the system optimized for this purpose, where the gRNA and the tracRNA have been fused. In the original system, which functions as an adaptive immune system in prokaryotes, that remembers past viral infections, the Cas9 endonuclease would associate with an individual gRNA and a tracRNA. Red, corresponding “guide RNA”; blue, “gRNA” target; broken black lines, protein or protein complex associated with the “gRNA”. Region of base paring between the gRNA and the target nucleic acid are indicated. Asterisks indicate the site of cleavage or modification in the target nucleic acid.
… the discovery of the first gRNAs […] predates the discovery of the CRISPR/Cas9 system by more than two decades.
In the following, I would like to give an account of when the term gRNA first appeared in the literature. Moreover, I will discuss how the concept itself has evolved and where its origins are. While I was not involved in any of the discoveries discussed here, I have some personal recollections relevant for the story. I was in close contact with Beat Blum, who sadly passed away three years ago, when his landmark paper appeared in which the term gRNA was christened (Blum et al, 1990). As the discovery of the first gRNAs by Beat nicely illustrates the intricate paths that often lie behind scientific discoveries, I made him the focus of my account.
The discovery of RNA editing
The story begins with Rob Benne at the University of Amsterdam who sequenced mitochondrial DNA fragments and the corresponding mRNAs of the parasitic protozoan Trypanosoma brucei. To his surprise, the genomic sequence encoding cytochrome oxidase subunit 2 (Cox2) and the sequence derived from the corresponding mRNA did not match, and he consistently found a four‐uridine insertion in the mRNA that was not coded for in the gene. The insertion corrected a frame shift predicted to occur in the Cox2 gene. After reluctantly accepting the unexpected results, Benne published his work in 1986 and postulated a process that he termed RNA editing, which precisely inserts four uridines at a defined position in the Cox2 transcript in order to produce a translatable mRNA (Benne et al, 1986).
His paper was initially greeted with skepticism, which eventually subsided when further, even more extreme cases of RNA editing were found, all in trypanosomatids. These editing events all consisted of multiple uridine insertions and/or deletions and were so extensive in some cases that the final mRNAs would not even hybridize to their genes anymore. RNA editing was now taken seriously by the broader scientific community and a number of review articles discussed its implications. The most perplexing question was where the information for the edited sequences comes from, since, despite extensive searches by several groups, a DNA or RNA template matching the edited sense or antisense sequences, could not be found in the mitochondrial genome of trypanosomes. Thus, some people thought that RNA editing would challenge the central dogma of Francis Crick, which led to some quite far‐fetched ideas of how the process may work.
… some people thought that RNA editing would challenge the central dogma of Francis Crick, which led to some quite far‐mission and he told everybody who wanted to work.
The very first guide RNAs
At that time, Beat Blum and I were PhD students, working with T. brucei in the laboratory of Thomas Seebeck at the University of Bern. Beat was interested in the newly discovered RNA editing process, and after graduating decided to do a postdoc with Larry Simpson at UCLA, one of the leading laboratories in the field. To that end, he secured a postdoc fellowship from the Swiss National Science Foundation (SNF). His proposal was very ambitious: like everybody working on RNA editing, Beat wanted to find out where the information that specifies RNA editing comes from. Unlike others, he refused to question the central dogma and was convinced that the problem could ultimately be reduced to specific base pairing with nucleic acids complementary to the edited sequences. In his SNF proposal, he meticulously outlined the various ways of how he wanted to find these postulated nucleic acids. In fact, even before he left for UCLA, finding this missing information for RNA editing became Beat's mission and he told everybody who wanted to hear it—and many who did not—that he will go to Larry Simpson’s laboratory and solve the problem of RNA editing once and for all. Not many people, including me, took him seriously at that time.
… RNA editing became Beat's mission and he told everybody who wanted to hear it – and many who did not – that he will [..] solve the problem of RNA editing once and for all.
Yet, it did not take long after Beat arrived in Larry’s laboratory before he struck gold. He did a computer analysis and found short sequences in the mitochondrial genome of the trypanosomatid Leishmania tarentolae that were very similar to short segments of the edited mRNAs and flanked on the 3' side by a short stretch that closely matched the non‐edited region of the same mRNA. Many people had tried the same approach before him, since the required sequence information had been available for a while, but everybody failed. What Beat did differently though was that he allowed G‐U base pairs to get the matches required for precise RNA editing. G‐U base pairs had been discovered a long time ago, and a review of their crucial importance for RNA function was published in the very first issue of EMBO Reports (Varani & McClain, 2000).
Beat subsequently showed that the sequences matching the edited regions were transcribed into short RNAs. Together with Larry Simpson as senior author and Norbert Bakalara, he published his results in January 1990 in Cell (Blum et al, 1990) and termed the newly discovered molecules “guide RNAs”. It is the first time this term appears in the literature. They also proposed a model by which the 5´ portions of gRNAs form hybrids with the immediate 3´ flanks of the pre‐edited regions of mRNA and thus direct the required editing activities to the correct site of the molecules. This is in essence what I refer to in this commentary as the gRNA concept.
It is worth mentioning that trypanosomal gRNAs were not only the first ones discovered, but they are still the most complex ones known to date. They bring the editing machinery—which contains endonuclease, terminal uridyltransferase, U‐specific exoribonuclease, and RNA ligase activities—to the correct site of the pre‐edited mRNA and instruct these enzymes to sequentially insert or delete the correct number of uridines (Fig. 1). We know today that RNA editing, the post‐transcriptional change of an mRNA coding sequence, is widespread and can be very diverse. However, RNA editing in the trypanosomatid mitochondrion is surprisingly still the only one known to rely on gRNAs.
SnoRNAs are guide RNAs
Six years after the discovery of the trypanosomal gRNAs, it became clear that small RNAs in the nucleolus, termed snoRNAs—which had been known for many years—also function as gRNAs. Zsuzsanna Kiss‐Laszlo discovered that snoRNAs guide the methylation of the 2’‐O‐hydroxyl position of defined nucleotides in rRNAs (Kiss‐Laszlo, Henry et al, 1996). One year later, the same group showed that a different set of snoRNAs mediates site‐specific conversion of uridines into pseudouridines in rRNA. A typical eukaryotic cell contains hundreds of different snoRNAs, many of which are encoded on introns. Together with proteins, which include the corresponding modification enzymes, these RNAs form ribonucleoprotein particles (RNPs) known as snoRNPs. Each snoRNA contains a stretch of nucleotides complementary to the sequence surrounding the nucleotide to be modified (Fig 1). This enables it to recognize the target RNA and to bring the modification enzymes to the correct physical location. The discovery that the snoRNAs are required for nucleotide modifications was unexpected, since methylation and pseudouridylation are common in bacterial rRNAs, but do not require guidance by small RNAs in these systems.
During the past decades, the function of more and more snoRNA‐like gRNAs has been determined. It is now clear that they also guide site‐specific modifications in small nuclear RNAs (snRNAs) involved in splicing, and in some cases of tRNAs. A subgroup of snoRNAs mediates the structural reorganization of rRNA prior to its processing and therefore has functions unrelated to nucleotide modifications. Furthermore, vertebrate telomerase contains a snoRNA‐type RNA subunit termed TERC that serves as a template for telomer elongation. Considering the great number of snoRNA‐like RNAs, with as yet unassigned functions found in eukaryotes, it is likely that even more novel roles for these molecules will be discovered in future.
Considering the great number of snoRNA‐like RNAs, with as yet unassigned functions found in eukaryotes, it is likely that even more novel roles for these molecules will be discovered in future.
Guide RNAs everywhere
After the pioneering discoveries made in the 1990s, a whole universe of novel small RNAs was discovered in 2000 and later. It turned out that these RNAs, the most common ones of which are the small interfering RNAs (siRNAs), microRNAs (miRNA), and the PIWI‐interacting RNAs (piRNAs), function as guide RNAs, which direct protein complexes to specific target RNAs (reviewed in Bartel, 2004; Czech, et al, 2018; Fig 1). siRNAs derive from long endogenous dsRNA, formed by sense and antisense transcripts, as well as from exogenously introduced dsRNAs of viral or synthetic origin. miRNAs and piRNAs are processed from larger precursors via different and complex maturations pathways. The maturation pathway of snRNAs and miRNAs produce short, double‐stranded RNAs (dsRNAs). Subsequently, the guiding strand of dsRNA intermediates that base pairs with the target RNA is incorporated into a protein complex containing a member of the argonaute protein family. Most argonaute proteins have endonuclease activity and, in the case of siRNAs which perfectly match their targets, cleave the substrate RNAs at the site determined by siRNAs. If the match is not perfect, as in the case of miRNAs that often hybridize to the 3´UTR of specific mRNAs, the result is translational repression or in some cases activation. Although piRNAs do not form double‐stranded intermediates, they associate with a subfamily of argonaute proteins. Their main function is to silence transposon activity in germline cells by transcriptional and post‐transcriptional mechanisms.
In summary, the small gRNA‐like RNAs play various roles in gene regulatory processes, the specificity of which is guaranteed by base pairing to their target RNAs. They can either guide degradation of their targets, affect their transcription or translation, or their epigenetic state by a number of different mechanisms.
Standing on the shoulder of giants
Was Beat, the discoverer of trypanosomal gRNAs, also the founder of the widely applicable gRNA concept? As it is mostly the case, fundamental discoveries can be traced back to multiple, incremental steps and are only rarely the revolutionary change they appear to be when first published. This also applies for the gRNA concept. The importance of RNA–RNA interactions for positioning enzymatic activities to specific sites on target RNAs had already been shown for mRNA splicing. This is a highly complex and dynamic process that involves numerous rearrangements of various snRNAs and their associated proteins. It requires two transesterification steps. Step 1 results in a free 5′ exon and an intron lariat − 3′ exon intermediate connected to the branch point, a conserved sequence within the intron. Step 2 subsequently leads to the joining of the 5′ exon with the 3′ exon. Both steps are ultimately catalyzed by the U6 snRNA. As early as in the 1980s, it was recognized that U1 snRNA, which forms the U1 snRNP with a number of proteins, shows complementarity to the 5´ splice site of mRNA precursors (Fig 2; Lerner, Boyle et al, 1980; Rogers & Wall, 1980). In 1986, it was shown that this initiates the splicing cycle (Zhuang & Weiner, 1986). Later in the splicing process, the branch point sequence is recognized by limited base paring with U2 snRNA, and the 5´ and 3´ splice sites are recognized again by U5 snRNA. The recognition of the intron boundaries is therefore to a large part the result of specific base pairing with various snRNAs, although proteins clearly contribute to it. Thus, it is fair to say that the early studies on the U1 snRNA (Lerner et al, 1980; Rogers & Wall, 1980; Zhuang & Weiner, 1986) mark the origin of the gRNA concept, even though the extent of complementary base pairings involved in the splicing reactions are generally less extensive than in the case of the smaller guide RNA‐like molecules. Beat and his co‐authors cite publications from the splicing field in their gRNA paper and likely were influenced by these studies.
Figure 2. Schematic depiction of mRNA splicing.
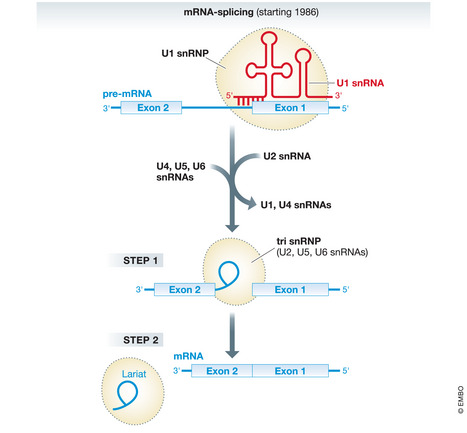
U1 snRNA base pairs with the 5´ intron exon junction of the pre‐mRNA and initiates the splicing cycle. The region of base paring between the U1 snRNA and pre‐mRNA is indicated. Next U2 snRNA recognizes the branchpoint sequence (not shown). Subsequently, more snRNAs and their associated proteins, including U6 snRNA that ultimately catalyzes the two step splicing reactions, assemble around the 5´ and 3´ splice sites, whereas other snRNAs dissociate from the pre‐mRNA. This results in a complex, termed tri snRNP, that contains U2, U5, and U6 snRNA. This complex catalyzes the first and the second splicing steps, resulting in 5′ exon and an intron lariat − 3′ exon intermediate before the 5´ and the 3´ exons are joined. There are many more, dynamic RNA–RNA interactions during the splicing cycle that are not shown. Red, U1 snRNA; blue, (pre‐)mRNA; broken black lines, snRNP complexes.
As it is mostly the case, fundamental discoveries can be traced back to multiple, incremental steps and are only rarely the revolutionary change they appear to be when first published.
Concluding remarks
Major breakthroughs are often achieved by relatively young scientists, who are less likely to be influenced by the dominant ways of thinking in a specific field and may therefore approach a problem without preconceptions. Beat was 36 years old when he discovered the gRNAs. However, in my opinion and quite ironically, it was not highly imaginative and innovative thinking that we associate with the young, which led to his breakthrough. Rather it was Beat’s stubborn and conservative attitude. He firmly believed, against much opposition in the field, that the problem of the missing information for RNA editing could ultimately be reduced to base pairing.
… in my opinion and quite ironically, it was not highly imaginative and innovative thinking that we associate with the young, which led to his breakthrough.
However, while thinking outside the box might not have been crucial, he needed an intimate knowledge of the box’s content and understood that RNA can also form G‐U base pairs. This insight was decisive to remove the annoying mismatches that prevented the precise alignment of gRNAs and edited mRNA sequences, and that had prompted many people to give up on looking for RNA templates to explain RNA editing.
We now know that RNA editing is widespread. It also occurs in plant mitochondria and chloroplasts, where it consists of site‐specific C‐to‐U conversion in many primary transcripts and can be very extensive. Would RNA editing first have been discovered in plants and not in trypanosomes, Beat Blum would probably have failed with his approach to the problem, since RNA editing in plants is not mediated by guide RNAs and base pairing. Instead, the site‐specific C‐to‐U conversions are specified by pentatricopeptide repeat proteins that can bind to RNA in a sequence‐specific way. Thus, as with many things in life, luck and being at the right time in the right place can help a lot to make a scientific discovery.
Acknowledgements
I would like to thank Sebastian Leidel and Oliver Mühlemann for critical comments on the manuscript. Research in my laboratory is supported by the NCCR "RNA & Disease" and by grant 175563 both funded by the Swiss National Science Foundation.
EMBO Reports (2020) 21: e51918.
References
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Benne R, Burg JVD, Brakenhoff JPJ, Sloof P, Boom JHV, Tromp MC (1986) Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819–826 [DOI] [PubMed] [Google Scholar]
- Blum B, Bakalara N, Simpson L (1990) A model for RNA editing in kinetoplastid mitochondria: "guide" RNA molecules from maxicircle DNA provide the edited information. Cell 60: 189–198 [DOI] [PubMed] [Google Scholar]
- Czech B, Munafo M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, Hannon GJ (2018) piRNA‐Guided Genome Defense: From Biogenesis to Silencing. Annu Rev Genet 52: 131–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss‐Laszlo Z, Henry Y, Bachellerie JP, Caizergues‐Ferrer M, Kiss T (1996) Site‐specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA (1980) Are snRNPs involved in splicing? Nature 283: 220–224 [DOI] [PubMed] [Google Scholar]
- Rogers J, Wall R (1980) A mechanism for RNA splicing. Proc Natl Acad Sci USA 77: 1877–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G, McClain WH (2000) The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep 1: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y, Weiner AM (1986) A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell 46: 827–835 [DOI] [PubMed] [Google Scholar]


