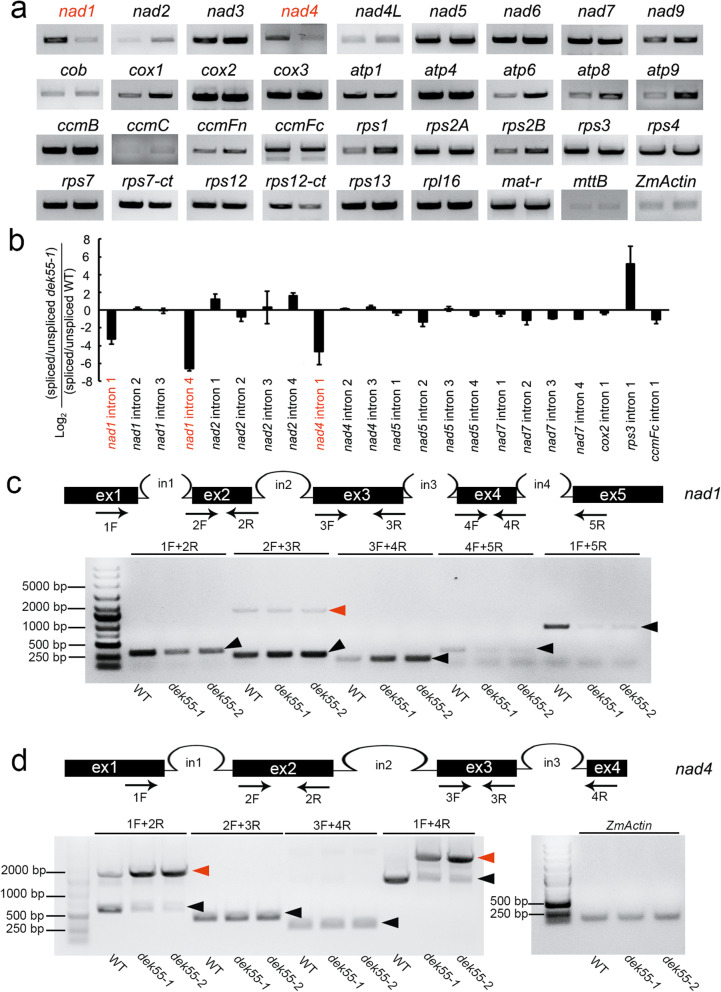
Fig. 5.
The posttranscriptional RNA processing of nad1 and nad4 was affected in dek55. a The expression of 35 mitochondrion-encoded genes in the WT (left) and dek55–1 (right) kernels was detected via RT-PCR. The ZmActin gene (GRMZM2G126010) was used as an internal control. Both nad1 and nad4 are marked in red because their transcript abundance significantly decreased. b The splicing efficiency of all 22 group II introns in maize mitochondrial-encoded genes was determined in dek55–1 and WT kernels via qRT-PCR. The values shown are the means of three biological replicates, and the error bars represent the standard deviations. c-d Schematic structure of the nad1 gene (c) and nad4 gene (d). The primers used for amplification are indicated. RT-PCR-based analysis of the intron splicing efficiency of nad1 in WT kernels and in dek55–1 and dek55–2 mutant kernels at 15 DAP. All the PCR products were confirmed by sequencing. The ZmActin gene (GRMZM2G126010) was used as an internal control. The unspliced and spliced fragments are indicated by red and black arrowheads, respectively. Exons are indicated as “ex”, and introns are indicated as “in”. The gel images in (a, c, d) were cropped; the original gel images are shown in Additional file 1: Figs. S2-S3