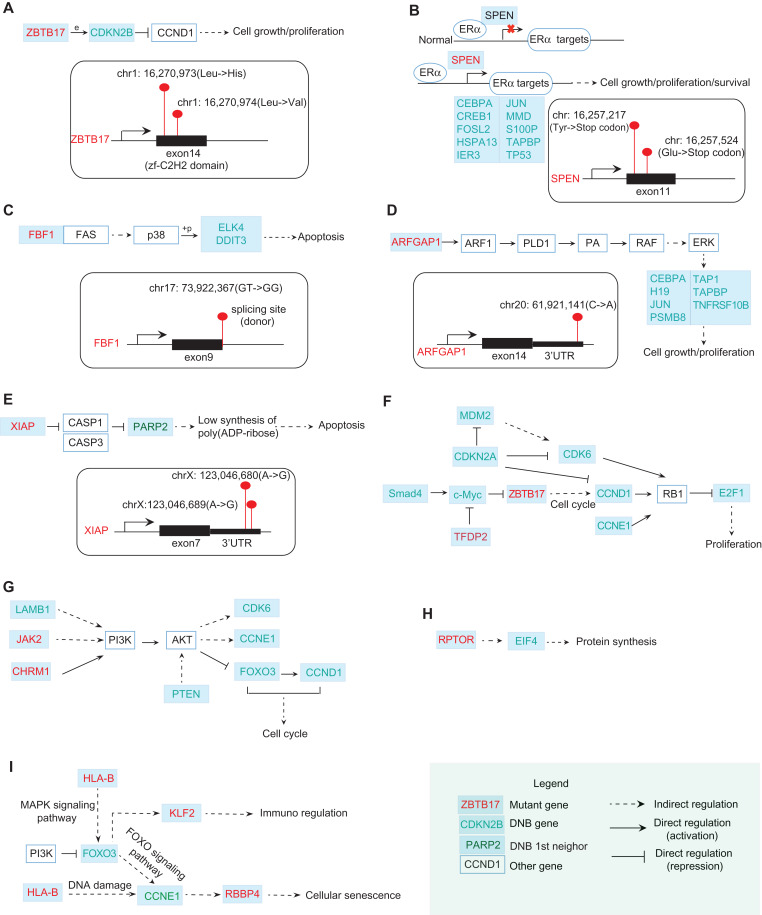
Figure 6.
The functional relations between mutation genes and DNB genes. (A) ZBTB17 with two missense mutations occurring in its zf-C2H2 domain can directly regulate the expression of a DNB gene CDKN2B. By directly repressing Cyclin D (CCND1), CDKN2B is involved in regulating cell cycle progression. (B) In normal breast cells, SPEN binds to ERα and negatively regulates the transcription of ERα targets. SPEN with nonsense mutations may re-activate ERα targets including DNB genes CEBPA, CREB1, FOSL2, etc. (C) A Fas-binding factor FBF1 has a mutation at the splicing site. Fas can induce the apoptosis by indirectly regulating some TFs. Among those TFs, two of DNB genes ELK4 and DDIT3 are involved. (D) ARFGAP1 with a mutation at 3′UTR may stimulate cell growth and proliferation by indirectly activating ERK for regulating some of DNB genes downstream, including CEBPA, H19, JUN, etc. (E) XIAP with two mutations at its 3′UTR encodes an X-linked inhibitor of apoptosis. One of the DNB neighbors at the first level, PARP2, is involved in this pathway. (F) From KEGG pathway in cancer, DNB genes SMAD4 and c-Myc regulate the mutant gene ZBTB17, which in turn affects DNB genes CCND1, E2F1 and thus induces the proliferation. (G) From PI3K–AKT signaling pathway, DNB genes LAMB1 and mutant genes JAK2, CHRM1 regulate the downstream DNB genes FOXO3, CCNE1, CDK6, etc. (H) The interplay between mutant gene RPTOR and DNB gene EIF4 takes part in the protein synthesis process. (I) The interplay between mutant genes HLA-B, RBBP4 and DNB genes FOXO3, CCNE1 is also involved in the cellular senescence pathway. The mutant genes above may interplay with DNBs to promote the development of drug resistance. In other words, the accumulated mutations eventually affect the DNB genes that subsequently cause the change of transcriptional landscape, enabling full-blown drug resistance.