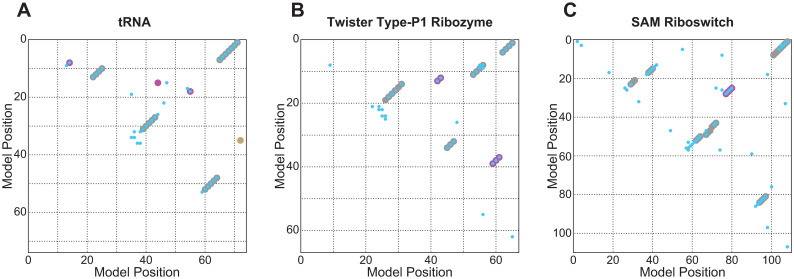
Fig 6. Gremlin-trained Potts models accurately predict 3D RNA contacts.
Top predicted 3D contacts from Gremlin-trained Potts models (cyan) against annotated nested base pairs (gray) and annotated tertiary contacts/pseudoknotted positions (pink) from the tRNA (A), Twister type P1 ribozyme (B), and SAM riboswitch (C) training alignments. For tRNA, annotated tertiary contacts are from a yeast tRNA-phe crystal structure [48]. A non-structural covariation between the tRNA discriminator base and the middle nucleotide in the anticodon (caused by aminoacyl-tRNA synthetase recognition preferences, not conserved base-pairing) is noted in orange [49].