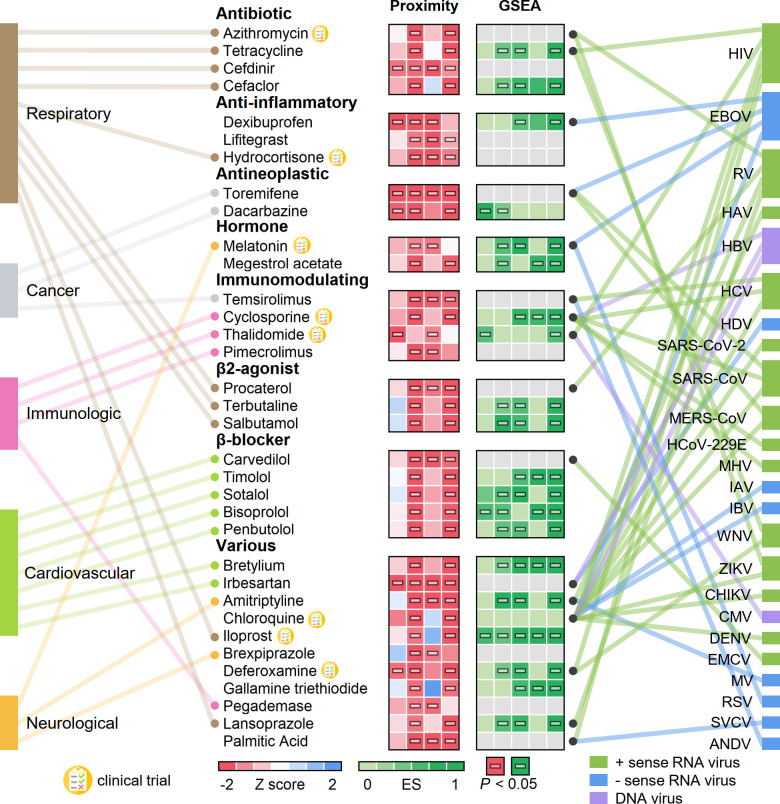
Fig 7. Network-based prediction of drug repurposing for COVID-19.
Thirty-four drugs from the top predicted list are highlighted, with the disease category they are approved for by the US Food and Drug Administration. We highlight 3 types of evidence: (1) network proximities of drugs’ targets across the 4 SARS-CoV-2 datasets (SARS2-DEG, SARS2-DEP, HCoV-PPI, and SARS2-PPI) in the human interactome, (2) gene set enrichment analysis (GSEA) scores across 5 coronavirus transcriptomics and proteomics datasets, and (3) literature-reported antivirus profiles. GSEA scores shown in grey indicate that these drugs cannot be evaluated due to the lack of data. Eight drugs that are currently being or have been tested in COVID-19 clinical trials are highlighted. Horizontal bars in boxes indicate P < 0.05. The data underlying this figure can be found in S6 Data. ANDV, Andes virus; CHIKV, chikungunya virus; CMV, cytomegalovirus; DENV, dengue virus; EBOV, Zaire ebolavirus; EMCV, encephalomyocarditis virus; ES, enrichment score; HAV, hepatitis A virus; HBV, hepatitis B virus; HCoV-229E, human coronavirus 229E; HCV, hepatitis C virus; HDV, hepatitis D virus; HIV, human immunodeficiency virus; IAV, influenza A virus; IBV, influenza B virus; MERS-CoV, Middle East respiratory syndrome coronavirus; MHV, mouse hepatitis virus; MV, measles virus; RSV, respiratory syncytial virus; RV, rhinovirus; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SVCV, spring viremia of carp virus; WNV, West Nile virus; ZIKV, Zika virus.