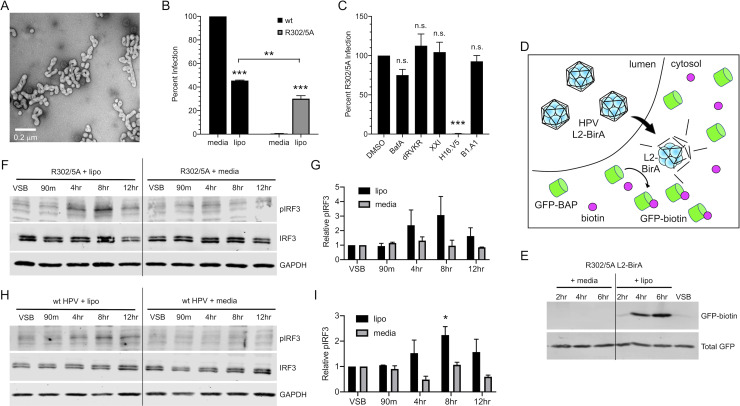
Fig 5. Bypassing HPV’s natural trafficking pathway activates cGAS/STING.
Addition of cationic lipids during infection with translocation-deficient R302/5A mutant HPV restores infectivity and allows for cGAS/STING sensing. (A) Electron micrograph of HPV complexed with the cationic lipid Lipofectamine 2000. (B) HaCaTs were infected with 2e8 vge/well of wildtype (capsid) or R302/5A virion +/- cationic lipid (lipo) for 4 hr and infection was measured by luciferase assay 24 hr post-infection. While naturally non-infectious, addition of cationic lipids restored infectivity of the R302/5A mutant to levels nearly comparable to those of wildtype HPV. Statistics calculated by one-way ANOVA (P < 0.001) followed by Tukey’s multiple comparisons test (**P < 0.01, ***P < 0.005), n = 3 biological replicates (C) R302/5A infection in the presence of cationic lipids is insensitive to biochemical inhibitors of endosomal acidification (BafA), furin (dRVKR), γ-secretase (XXI), and BPV1-specific Nab B1.A1, but is sensitive to HPV16-specific H16.V5. With exception of H16.V5 (***P < 0.005), differences were not significant by one-way ANOVA followed by Dunnett’s multiple comparisons test, n = 3 biological replicates. (D) Schematic of the membrane penetration assay using L2-BirA viruses in HaCaT cells stably expressing cytosolic GFP-BAP. Conditions enabling early membrane penetration result in L2-BirA dependent generation of GFP-biotin. (E) Membrane penetration assay. R302/5A L2-BirA virions (250 ng L1) were complexed +/-lipo or media as described. HaCaT-GFP-BAP cells were infected with virus/complexes for the indicated times prior to detection of GFP-biotin and total GFP by SDS-PAGE and western blot. (F-I) Cells were infected for 4 hr with R302/5A (F, G) or wildtype HPV16 (H, I) virion equivalent of 250 ng DNA +/- cationic lipids (lipo) as described in Materials and Methods. Addition of cationic lipids during virus infection resulted in IRF3 phosphorylation at 4 hr and 8 hr, while virus infection in media alone did not induce IRF3 phosphorylation, representative blots shown of n = 2 biological replicates in both (F, H). (G, I) Densitometric quantification of western blots. (G) Statistics were calculated by two-way ANOVA followed by Dunnett’s multiple comparisons test. Due to technical aspects of background variability and few replicates, differences did not reach statistical significance (Pinteraction = 0.331, P = 0.125 at 8 hr time point), n = 2 biological replicates. (I) Statistics were calculated by two-way ANOVA (Pinteraction = 0.1359), followed by Dunnett’s multiple comparisons test (*P < 0.05), n = 2 biological replicates.