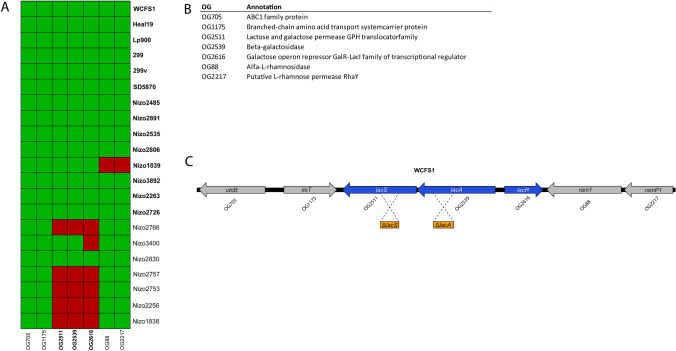
Figure 3.
Gene cluster, identified in L. plantarum strains that readily utilize GOS. (A) Heatmap displaying the in silico gene-trait matching results based on the association between presence (green) or absence (red) of specific OGs and GOS utilization phenotypes “A” (strain names in bold) and “B” (strain names in normal font). (B) Predicted functions of OGs encoded in the identified operon and flanking genes in reference strain WCFS1. (C) Gene map of the lac operon and flanking genes in reference strain WCFS1 with OGs identified by gene-trait matching (blue), and genes flanking the operon (grey). The lac operon and flanking regions of WCFS1 are conserved in L. plantarum strain NC8, and homologous regions cloned in suicide vector pNZ5319 for disruption of candidate genes by single cross-over events in NC8 are presented in orange boxes for lacS (ΔlacS) and lacA (ΔlacA).