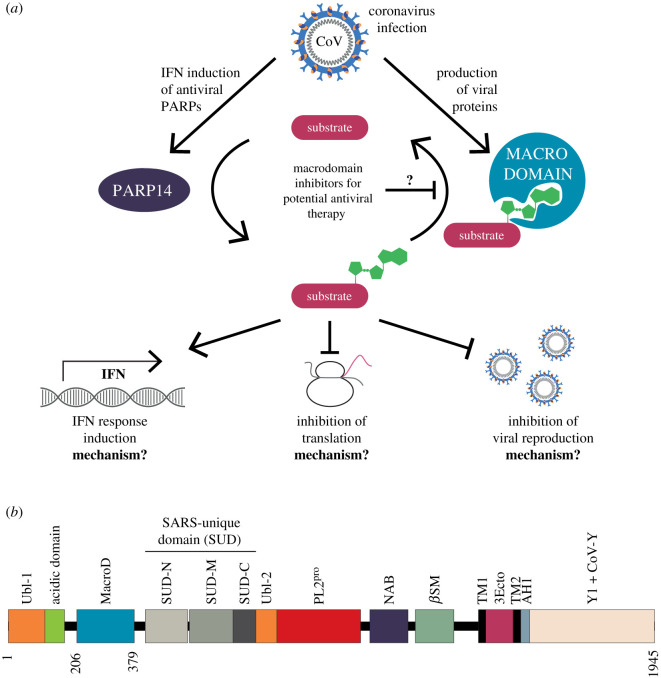
Figure 1.
Model for the role of ADP-ribosylation following coronavirus infection. (a) Infection of cells with coronavirus leads to the induction of an interferon (IFN) response and the accumulation of human antiviral as well as viral proteins, such as human PARP14 and viral macrodomain, respectively. The ADP-ribosylation activity of PARP14 stimulates maintenance of IFN responsive gene expression, downregulation of translation and prevents viral replication. This is antagonised by the viral macrodomain, part of the multidomain non-structural protein 3 (nsp3), which exhibits (ADP-ribosyl)hydrolase activity and is required for evasion of immune responses and efficient viral replication. However, both the mechanism underlying the changes in cellular processes as well as the precise targets for (de)ADP-ribosylation are currently unknown. (b) Schematic overview of the nsp3 domain architecture of SARS-CoV-2. Ubl, ubiquitin-like domain; MacroD, macrodomain of the MacroD-like class; SUD, SARS-unique domain (structurally subdivided into N-terminal, middle and C-terminal SUD with N- and M-SUD harbouring a macrodomain fold and SUD-C a ‘N-terminal domain of CyaY-like' fold); PL2pro, papain-like protease 2; NAB, nucleic acid binding domain; βSM, betacoronavirus-specific marker; TM, transmembrane region; 3Ecto, Nsp3 ectodomain (also termed ‘zinc-finger domain'); AH1, aliphatic helix 1; Y1 + Y-CoV, C-terminal region of unknown function with the initial domain (Y1) widely conserved and an apparently coronavirus specific (Y-CoV) addition. Domain boundaries were inferred by sequence comparison following a summary by Lei and colleagues [6].