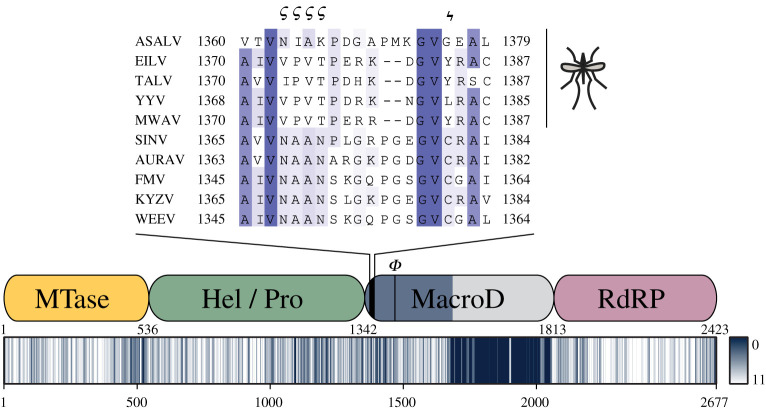
Figure 6.
Loss of catalytic residues in the macrodomain of insect-restricted alphaviruses. Non-structural polyprotein (precursors of nsp1–4) sequences were aligned using Mafft L-INS-I and position-specific physico-chemical properties conservation scores plotted as heat map (lower panel; alignment position are indicated below the plot). Positions of nsp1–4 (nsp1, yellow; nsp2, green; nsp3 blue; nsp4, red) are indicated in the middle panel and inferred cleavage site positions for Agua Salud alphavirus (ASALV) given below the scheme. Names of the primary domains residing within these proteins are abbreviated as follows: MTase, guanine-7-methyltransferase (also possesses guanylyl-transferase [GTase] activity); Hel, helicase; Pro, protease; RdRP, RNA-dependent RNA polymerase. ϕ indicates the position of the aromatic residue important for distal ribose positioning in vertebra-infecting alphaviruses. This residue is substituted by lysine or glutamine in insect-restricted alphaviruses. The poor alignment and low conservation score in the C-terminal region of nsp3 (indicated in grey) is due to the presence of a hypervariable domain within this region. The alignment for the catalytic loop region of the macrodomain is given in the top panel and NAAN motif (ϛ) and proposed catalytic cysteine (ϟ) indicated above. The full alignment is given in electronic supplementary material, figure S4.