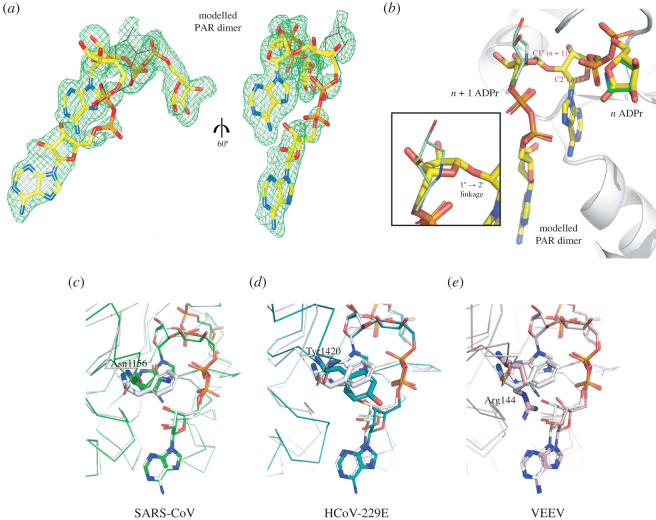
Figure 8.
Two ADP-HPD ligands bound to S2-MacroD mimic PAR dimer binding. (a) Modelled PAR dimer (yellow sticks) placed in the composite omit map (countered at 2σ) of the two ligands ADP-HPD from S2-MacroD:ADP-HPD structure (orientation as figure 3c,d). Black lines trace the original position of the ADP-HPD ligand. (b) Close up of the overlay between the model PAR dimer (yellow) and the two ADP-HPD ligands (n, canonical ADP-HPD [dark green]; n + 1, non-canonical ADP-HPD [light green]) bound to S2-MacroD (white). The framed inset is a zoomed view of the distal ribose of the n + 1 ADPr highlighting the twist respect to the pyrrolidine ring of the second ADP-HPD ligand. (c–e) Energy minimized models of PAR dimer in selected viral macrodomains. The models were generated using (c) SARS-CoV:ADPr (PDB 2FAV), (d) HCoV-229E:ADPr (PDB 3EWR) and (e) VEEV:ADPr (PDB 3GQO) as initial structures and overlaid to S2-MacroD-PAR dimer model in white. Amino acids corresponding to Phe360 of S2-MacroD structure are shown in sticks and are Asn1156 (c), Tyr1420 (d) and Arg144 (e) with numbering corresponding to alignment in electronic supplementary material, figure S2.