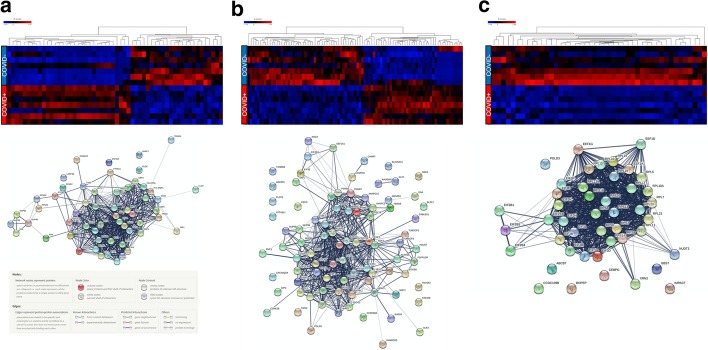
Fig. 3.
Gene expression and predicted protein–protein interaction networks of transcriptional differences in circulating leukocytes from COVID19 + and COVID19− critically ill patients. Differentially expressed genes (1311) were submitted to the Reactome Pathway Browser, resulting in the identification of three major pathway classifications (shown as heat maps, left to right): a interferon (interferon signalling, interferon alpha/beta signalling, anti-viral mechanism by IFN-stimulated genes); b cell cycle regulation (cell cycle, mitotic, G1/S transition, mitotic G1 phase and G1/S transition, G1/S-specific transcription); and c protein translation/ribosomes (GTP hydrolysis and joining of the 60S ribosomal subunit, formation of a pool of free 40S subunits, formation of the ternary complex, the 43S complex, eukaryotic translation initiation, Cap-dependent translation initiation, L13a-mediated translational silencing of ceruloplasmin expression, eukaryotic translation elongation, peptide chain elongation, translation initiation complex formation, response of EIF2AK4 (GCN2) to amino acid deficiency). The gene lists corresponding to these three pathway classifications were submitted to String-db (https://string-db.org/cgi/input.pl) to predict the protein–protein interaction networks (shown below each corresponding heat map)