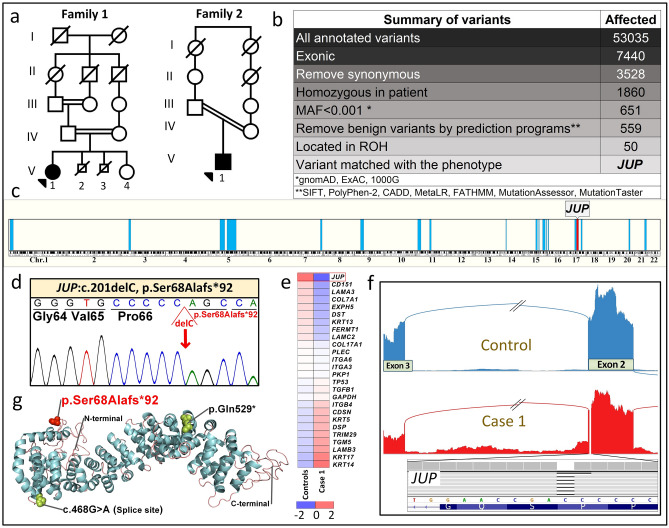
Figure 3.
Identification of biallelic JUP mutations. (a) Pedigrees of two families with consanguinity. The proband in each family is indicated by an arrowhead. (b) Identification of the mutation in JUP by analysis of annotated variants from RNA-Seq data in Patient 2 using bioinformatics filtering steps indicated. (c) Homozygosity mapping based on RNA-Seq data localized the JUP gene within a homozygosity block of 10.7 Mb on chromosome 17. (d) Sanger sequencing confirmed the presence of the previously unreported homozygous mutation, JUP: NM_002230: exon 2, c.201_201delC, p.Ser68Alafs*92, in both probands. (e) Heatmap visualization of transcriptome analysis revealed markedly reduced level of JUP expression in comparison to the pool of six healthy controls when compared to other genes associated with EB phenotypes and randomly selected housekeeping genes. (f) Sashimi plot of the transcriptome profile of the mutant JUP mRNA revealed normal splicing, indicating lack of exon skipping and/or significant intron retention. (g) Protein visualization of full-length JUP consisting of 745 amino acids. The mutation identified in this study (red) is predicted to result in a frameshift and early truncation of the protein. Two previously reported JUP mutations in patients with similar phenotype are shown (black). The figure was rendered in PyMol (v.2, Schrodinger, New York, NY). For details of WES, homozygosity mapping, RNA sequencing and protein modeling, see PyMol, http://Rymol.org18,24,28,43.