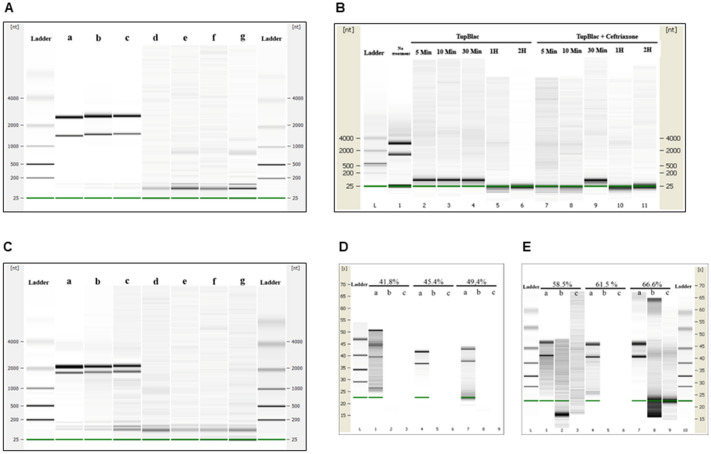
Figure 5.
Digital gel images of RNase activity of expressed Tupanvirus protein TupBlac on RNAs originating from E. coli, Acanthamoeba castellanii, and bacteria that differ by the G + C-content of their genome. RNA samples (1 µg) were incubated with 15 µg of TupBlac at 30 °C in the absence or presence of 10 µg/mL of sulbactam or 200 µM of ceftriaxone. Nuclease activity was visualized as digital gel images performed using the Agilent Bioanalyzer 2100 with the RNA 6000 Pico LabChip (Agilent Technologies, Palo Alto, CA). Each part A to E of the figure correspond to different digital gel images generated by the Agilent Bioanalyzer. (A) RNA used as substrate was from Escherichia coli; no treatment (a); buffer (b); blank control made with bacterial lysates purified by the same process and collected as Tupanvirus beta-lactamase (c); TupBlac in the absence (d) or presence (e) of 10 µg/mL of sulbactam, of 200 µM of ceftriaxone (f), or of 10 mM of EDTA (g). (B) RNA used as substrate was from E. coli; reactions were stopped at different times (5 min, 10 min, 30 min, 1 h and 2 h) by the addition of proteinase K (10 µg) and incubation for 1 h at 37 °C. The first lane corresponds to no treatment; lanes 2 to 6 correspond to RNA treatment with TupBlac in the absence of ceftriaxone; lanes 7 to 11 correspond to RNA treatment with TupBlac in the presence of ceftriaxone. (C) Nuclease activity on RNAs originating from Acanthamoeba castellanii; no treatment (a); buffer (b); blank control (c); TupBlac in the absence (d) or presence (e) of sulbactam (10 µg/mL), ceftriaxone (200 µM) (f), or EDTA (10 mM) (g). (D,E) nuclease activity on RNAs originating from bacteria that differ by the G + C-content of their genome, as indicated at the top of the digital gel image. For each RNA, three samples were analyzed: no treatment (a); treatment with TupBlac in the absence of sulbactam (b); and treatment with TupBlac in the presence of sulbactam (c).