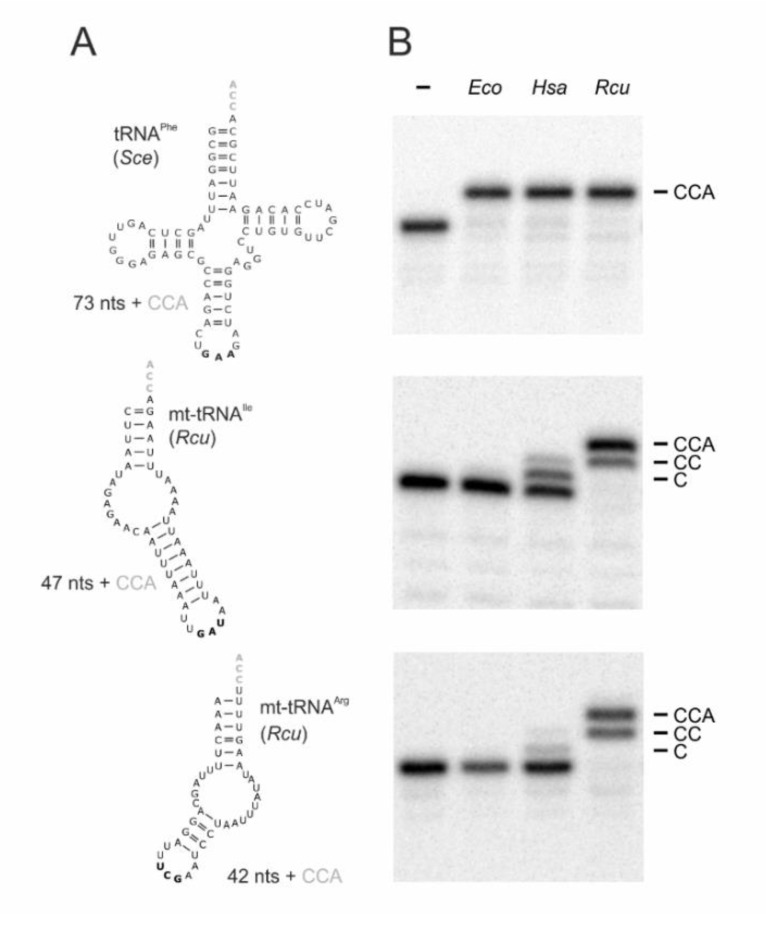
Figure 2.
CCA-addition on conventional and hairpin-like tRNA substrates. (A) tRNAPhe from Saccharomyces cerevisiae (Sce) represents a conventionally structured tRNA substrate of standard size (73 nts without CCA), while the mitochondrial tRNAs for isoleucine and arginine from R. culicivorax (Rcu) considerably deviate in size (47 and 42 nts, respectively; both without 3′-terminal CCA-triplet) and structure, lacking both D- and T-arms. Anticodons are indicated in bold. (B) CCA-addition on radioactively labeled tRNA transcripts catalyzed by the corresponding enzymes (20 ng each) of Escherichia coli (Eco), H. sapiens (Hsa), and R. culicivorax (Rcu). Incubation without enzymes represent negative controls (−). All enzymes completely convert the canonical tRNAPhe from S. cerevisiae into a mature transcript with CCA-end. On armless mt-tRNAs, the E. coli enzyme shows no activity at all, while the human enzyme adds only one or two C residues to mt-tRNAArg and mt-tRNAIle, respectively. In contrast, the enzyme of R. culicivorax readily synthesizes a complete CCA-end on both transcripts, although the time of incubation was not sufficient for 100% A-addition. The experiment was done in three independent replicates. The panel shows a representative autoradiogram.