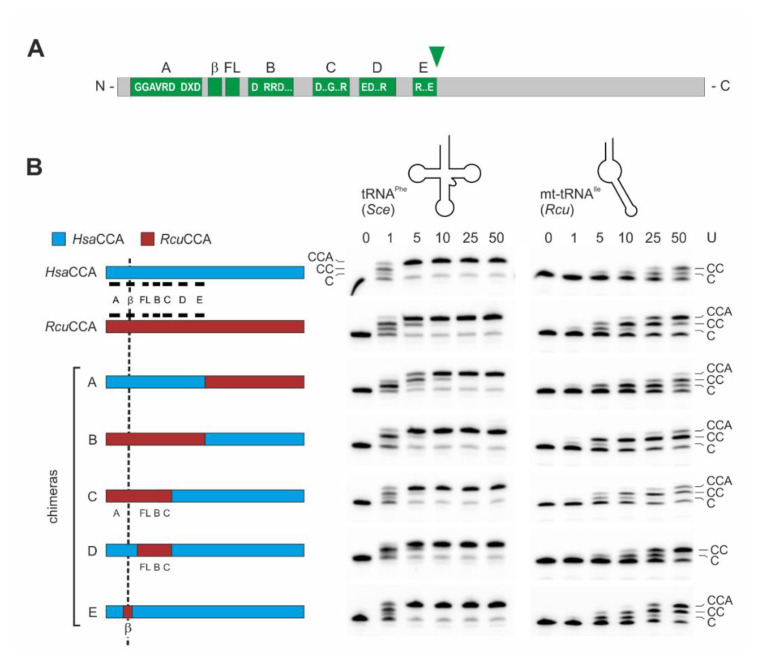
Figure 3.
Catalytic activity of wild-type and chimeric enzymes on tRNAPhe and mt-tRNAIle. (A) Bar representation of class II CCA-adding enzymes. The N-terminal region contains the catalytic core consisting of five motifs A to E, a β-turn element (β) and a flexible loop (FL). The green arrowhead indicates the fusion position for enzyme chimeras A and B. (B) Left: Bar representation of tested enzyme chimeras consisting of HsaCCA (cyan) and RcuCCA (red) regions. Catalytic core elements are indicated in black. The reciprocal chimeras A and B are fused after position E213, downstream of motif E. The replaced β-turn element (β) in chimera E is located between motif A and the flexible loop (FL). Gel panels: CCA-addition on the conventional tRNAPhe from yeast (Sce, left) and the armless mt-tRNAIle from R. culicivorax (Rcu, right) with increasing amounts of enzymes indicated as arbitrary units (U). All enzymes catalyze an efficient CCA-incorporation on the conventional tRNA. On the armless tRNAIle, the Romanomermis wt enzyme synthesizes a complete CCA-end, while the corresponding human enzyme adds only two C-residues. Chimeras A and B also add the terminal A, but at a somewhat reduced level. On this tRNA substrate, chimera B, carrying the catalytic core of RcuCCA, is more efficient than chimera A, where A-incorporation is only visible at the highest enzyme concentration. Chimera C still shows full CCA-addition, whereas no terminal A-incorporation is observed for chimera D. Chimera E shows an efficiency comparable to that of the RcuCCA wt enzyme, indicating the importance of the β-turn region in the reaction on armless tRNAs. The fact that chimera E is more active than chimera B likely reflects differences in the compatibility of the chosen fusion positions in these chimeras, resulting in different protein folding and/or catalytic efficiency. For each construct, up to four independent experiments were performed. For each tRNA substrate, a representative gel is shown.