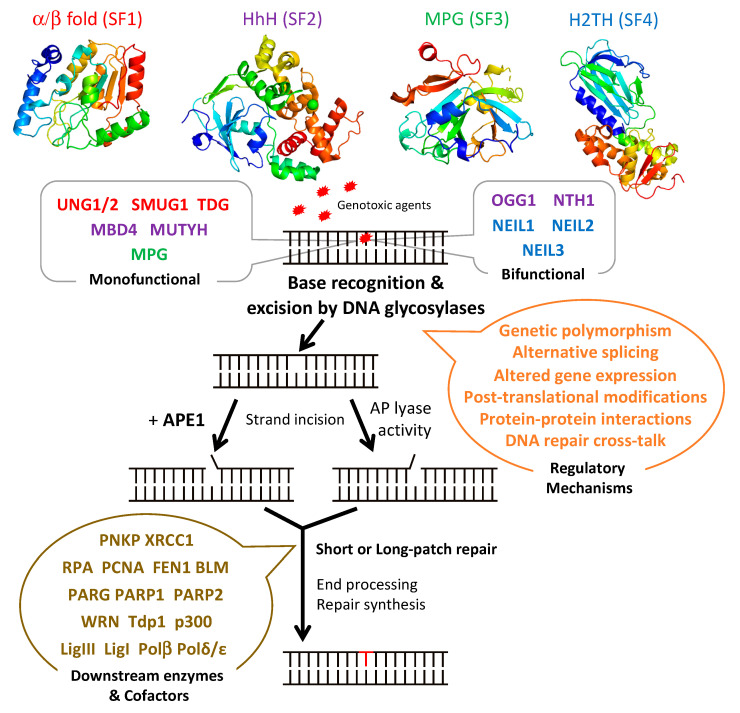
Figure 1.
Schematic diagram illustrating the steps and enzymes involved in the base excision repair (BER) pathway. Representative structures of the different superfamilies (SF) of DNA glycosylases (SF1, α/β fold family, red; SF2, helix-hairpin-helix (HhH) family, purple; SF3, 3-methyl-purine glycosylase (MPG) family, green; SF4, helix-two-turn-helix (H2TH) family, blue) responsible for recognition and removal of damaged bases are shown. After cleavage of the damaged strand by an apurinic/apyrimidinic (AP) endonuclease, APE1, or by the AP lyase activity of bifunctional DNA glycosylases, downstream BER enzymes together with several cofactors (listed in brown) prepare the damaged site for de novo synthesis using one of two sub-pathways: short-patch or long-patch repair. DNA glycosylases are tightly regulated at the gene, mRNA, and protein levels by a set of regulatory systems (listed in orange). UNG1/2: uracil-N glycosylase 1 or 2; SMUG1: single-strand-specific monofunctional uracil DNA glycosylase 1; TDG: thymine DNA glycosylase; MBD4: methyl-CpG-binding protein 4; MUTYH: MutY homolog DNA glycosylase; OGG1: 8-oxo-G DNA glycosylase 1; NTH1: endonuclease III-like 1; NEIL1-3: endonuclease VIII-like 1-3.