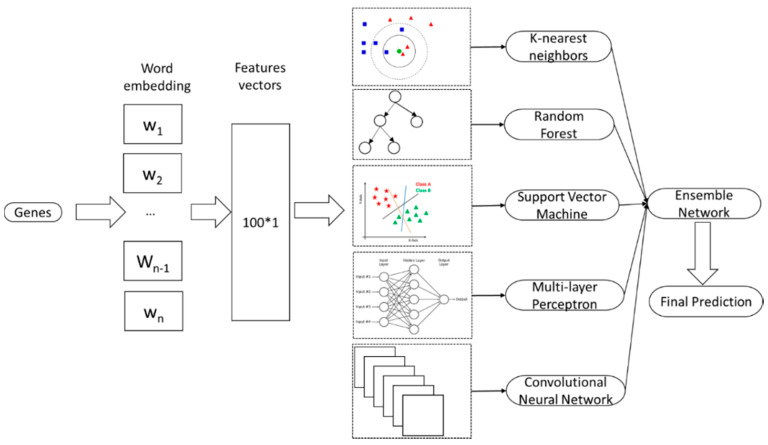
Figure 3.
Work flow of the study in identifying essential genes using sequence information. The input was comprised of genes with different lengths and containing different nucleotides. The word-embedding features were extracted by using the fastText package and then learnt by an ensemble deep neural network. After the ensemble network, the output contained binary probabilities to show whether the represented genes belonged to essential genes. Red, green triangles, green circle, blue squares and red pentagons are examples of data points.