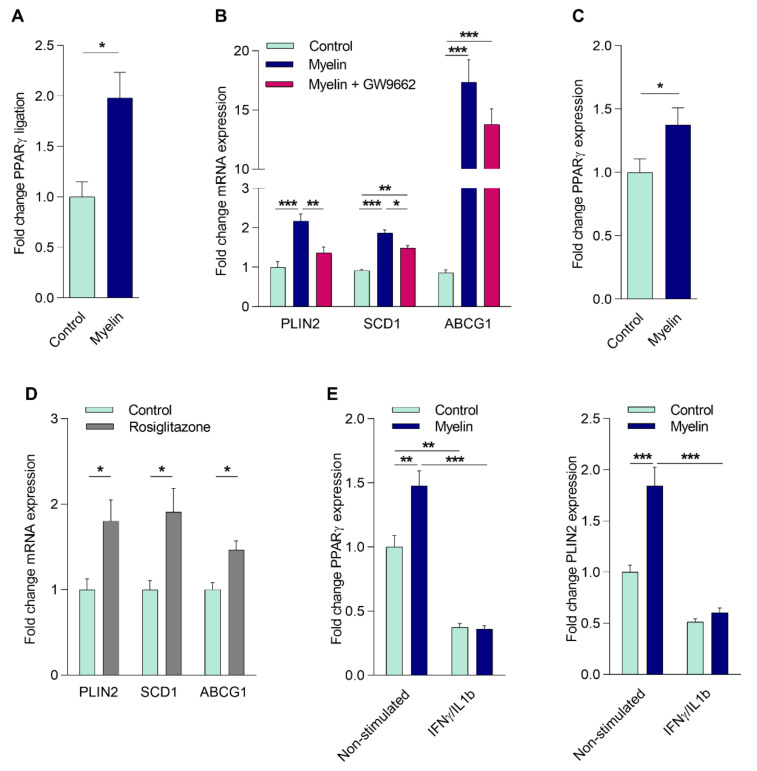
Figure 2.
Myelin uptake activates PPARγ in phagocytes. (A) CHME3 cells treated with myelin or left untreated. PPARγ ligand-binding activity was determined using the GAL-4-PPARγ chimera assay system (n = 5). (B) mRNA expression of PPARγ response genes in MDMs from healthy controls (HCs) stimulated with myelin in the presence or absence of GW9662 (25 µM; n = 8). (C) PPARγ mRNA expression in MDMs from HCs treated with myelin compared to non-stimulated cells (n = 11). (D) mRNA expression of PPARγ response genes in MDMs from HCs stimulated with rosiglitazone (1 µM; n = 9). (E) PPARγ and perilipin 2 (PLIN2) expression in MDMs from HCs treated with IFNγ/IL1β, followed with or without stimulation with myelin (n = 6). Values represent the mean ± S.E.M. Statistical significance (A,C,D; Mann–Whitney test, B; one-way ANOVA with Tukey’s multiple comparison correction, and E; two-way ANOVA with Sidak’s multiple comparison correction) is indicated with asterisks: * p ≤ 0.05, ** p ≤ 0.01, and *** p ≤ 0.001. n represents the total number of biological replicates (A) or donors (B–E) included in the experiment.