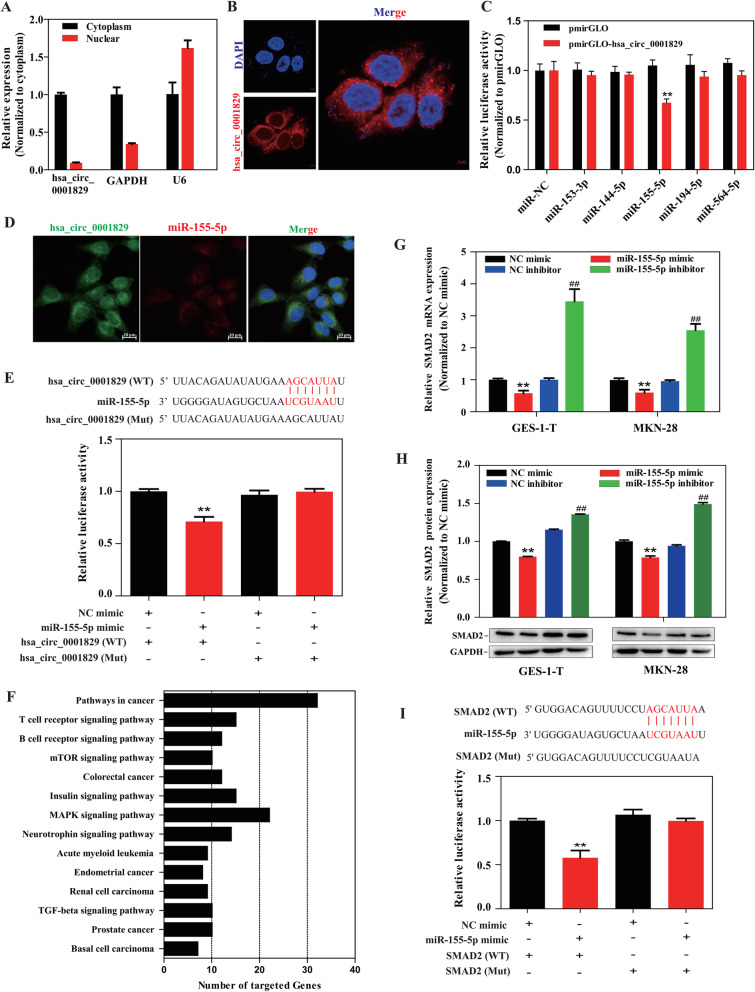
Fig. 5.
hsa_circ_0001829 acts as a molecular sponge for miR-155-5p, and SMAD2 was validated as a target gene of miR-155-5p. a Cytoplasmic and nuclear distribution of hsa_circ_0001829 in GES-1-T cells was detected by qRT-PCR. b The subcellular localization of hsa_circ_0001829 in GES-1-T cells by FISH assay. The nuclei were stained with DAPI for blue color, and the cytoplasmic hsa_circ_0001829 was stained for red color. c Luciferase activity in GES-1-T cells co-transfected with five miRNA mimics and hsa_circ_0001829 wild-type luciferase reporter vector. d Dual RNA FISH for hsa_circ_0001829 (green) and miR-155-5p (red) was detected in GES-1-T cells, miR-155 was co-localized with hsa_circ_0001829 in cytoplasm, with nuclei staining with DAPI (blue). Scale bars: 20 μm. e (Up) The binding sites of wild type or mutant hsa_circ_0001829 with miR-155-5p. (Down) Dual luciferase reporter assays demonstrated that hsa_circ_0001829 could sponge miR-155-5p. f KEGG pathway analysis of the 556 predicted target genes. g The effects of miR-155-5p mimics and inhibitors on the expression of SMAD2 mRNA detected by qRT-PCR. *vs NC mimic group, #vs NC inhibitor group. h The effects of miR-155-5p mimics and inhibitors on the protein expression level of SMAD2 by western blot. *vs NC mimic group, #vs NC inhibitor group. i (Up) The binding sites of wild type or mutant SMAD2 3′-UTR with miR-155-5p. (Down) Dual luciferase reporter assays demonstrated that SMAD2 is a direct target of miR-155-5p. *p < 0.05, **p < 0.01, # p < 0.05, ## p < 0.01