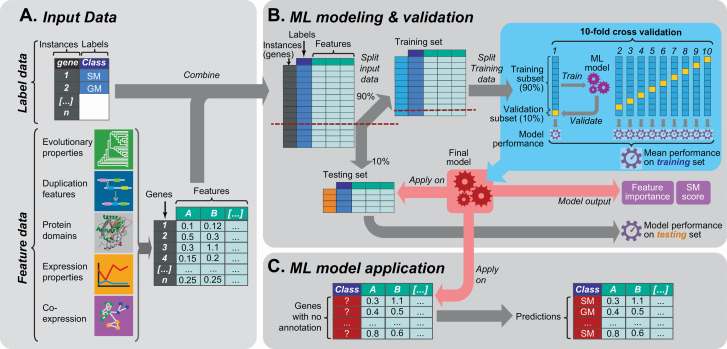
Figure 1.
Machine learning workflow used in this study. (A) Schematic showing the input data for machine learning. The first inputs are labelled instances, collectively referred to as the model training set. In this case the instances are genes and the labels are the gene classes (response variable; either specialized or general metabolism, SM or GM). The second input is features, or the predictive variables in the model. In this study, five feature categories, which each contain multiple features, were utilized: evolutionary properties, duplication features, protein domains, expression properties and co-expression data. Each gene (instance) has a value for each feature. (B) The machine learning process. First the data set was split into training (90 %) and testing (10 %) sets. Next, equal numbers of training instances (i.e. 500 GM and 500 SM genes) were randomly selected from the training set to learn prediction models. This step was repeated 100 times, with different subsets of GM/SM genes selected from the training set in each repeat, to assess the robustness of prediction models. For each repeat, a 10-fold cross-validation was performed where the selected instances were further divided into a training subset (90 %) for building the model and a cross-validation subset (10 %; distinct from the testing set withheld from model building) to evaluate the model. After cross-validation, the optimal parameters were chosen to establish the final model for a given training/feature data set. Model performance assessed using the cross-validation sets was represented using the average F-measure of all repetitions. In addition to assessing performance based on cross-validation, another F-measure was calculated for the final model based on its application to the testing set that was held out from the beginning and never used for training. (C) The final model is applied on unannotated enzymatic genes to make predictions.