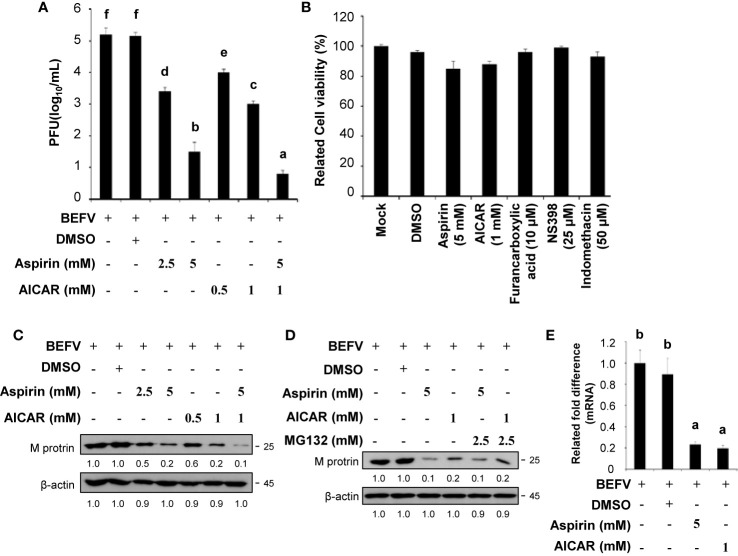
Figure 1.
Aspirin and 5-aminoimidazole-4-carboxamide-1-β-riboside (AICAR) inhibit viral growth. (A) Madin-Darby bovine kidney (MDBK) cells were pretreated with or without aspirin (2.5 and 5 mM) or AICAR (0.5 and 1 mM), respectively, for 30 min and then infected with BEFV at an MOI of 1 for 18 h. The effect of aspirin and AICAR on BEFV production was determined. Significance between the treatments was determined by Duncan’s Multiple Range Test (MDRT) using SPSS software (Version 20.0). Means with common alphabets (a, b, c, d,e, f) denotes no significance at p <0.05. Each value represents mean ± SE of three independent experiments. (B) To examine whether the compounds used in this study had the deleterious effects on cells, cell viability was determined using the MTT assay. Each value represents mean ± SE of three independent experiments. (C, D) The levels of the BEFV M protein in aspirin- and AICAR-treated MDBK cells were examined (C) in the presence or absence of proteasome inhibitor MG132 (D). The levels of indicated proteins in the BEFV-infected group were considered onefold. The inactivation folds indicated below each lane were normalized against values for the BEFV-infected group. Protein levels were normalized to those for β‐actin. Signals in all Western blots were quantified with ImageJ software. All experiments were conducted in three independent experiments. (E) The BEFV M and GADPH mRNA levels were quantified by real-time qRT-PCR in BEFV-infected MDBK cells in the presence or absence of indicated drugs. In real-time qRT-PCR amplification of the M and GADPH genes, MDBK cells were infected with BEFV at an MOI of 1. The BEFV-infected cells were collected at either 24 hpi, and total RNAs were extracted for real-time qRT-PCR. Significance between the treatments was determined by Duncan’s Multiple Range Test (MDRT) using SPSS software (Version 20.0). Means with common alphabets (a, b) denotes no significance at p <0.05. Each value represents mean ± SE of three independent experiments.