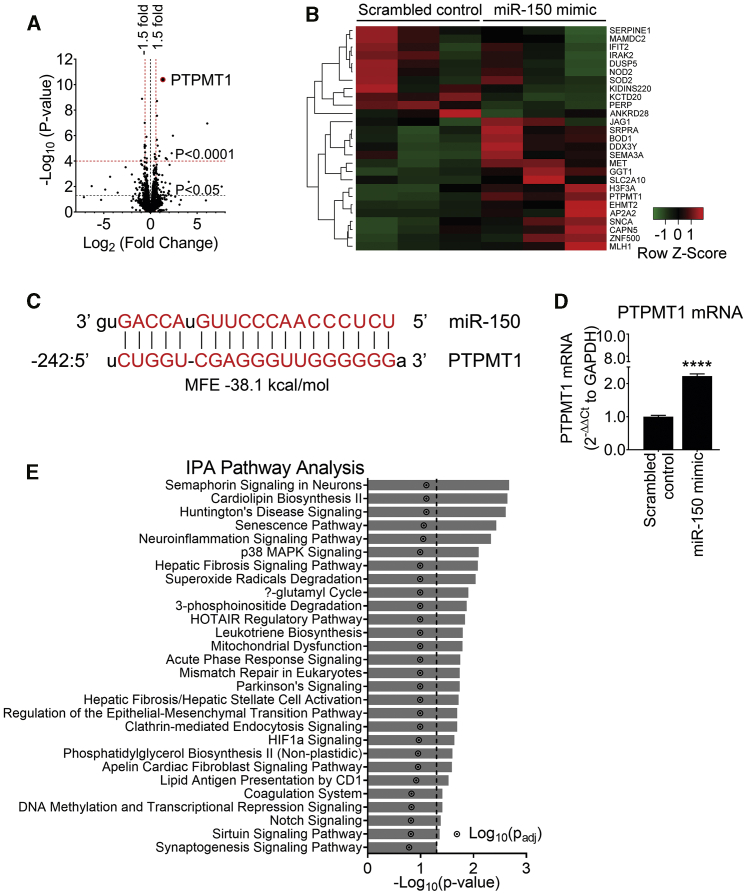
Figure 3.
RNA sequencing analysis in miR-150-overexpressing HPAECs shows PTPMT1 as the most upregulated gene
To identify miR-150 signaling mediators, HPAECs from three different donors were transfected with miR-150 or scrambled control (20 nM) and RNA was extracted for RNA sequencing in three independent experiments. (A) Volcano plot of differentially expressed genes (DEGs). Each point represents the difference in expression (log2 fold difference) between groups and the associated significance of this change (independent unpaired sample t test). PTPMT1 is highlighted as the most upregulated gene (fold change of 2.51, p = 3.98 × 10−11, adjusted p [p-adj] value = 5.48 × 10−7; n = 3). (B) Heatmap showing 26 most significant genes after multiple test correction using the Benjamini-Hochberg procedure (p-adj < 0.05). Green and red represent downregulation and upregulation, respectively. (C) miR-150 predicted binding sequence with the top minimum free energy (MFE) event (−31.3 kcal/mol) in the promoter (242 bp 5′ upstream) region of the PTPMT1 gene (chr11:47565430–47573461) using RNAhybrid. (D) PTPMT1 mRNA levels in cells transfected with control miRNA. Bars are mean fold changes of normoxia control ± SEM. n = 5. ∗∗∗∗p < 0.0001, comparisons with normoxia control (by unpaired Student’s t test). (E) Significantly enriched pathways (p < 0.05) regulated by miR-150; Ingenuity Pathway Analysis (IPA; version 01-12) of top 26 differentially expressed genes in cells transfected with miR-150.