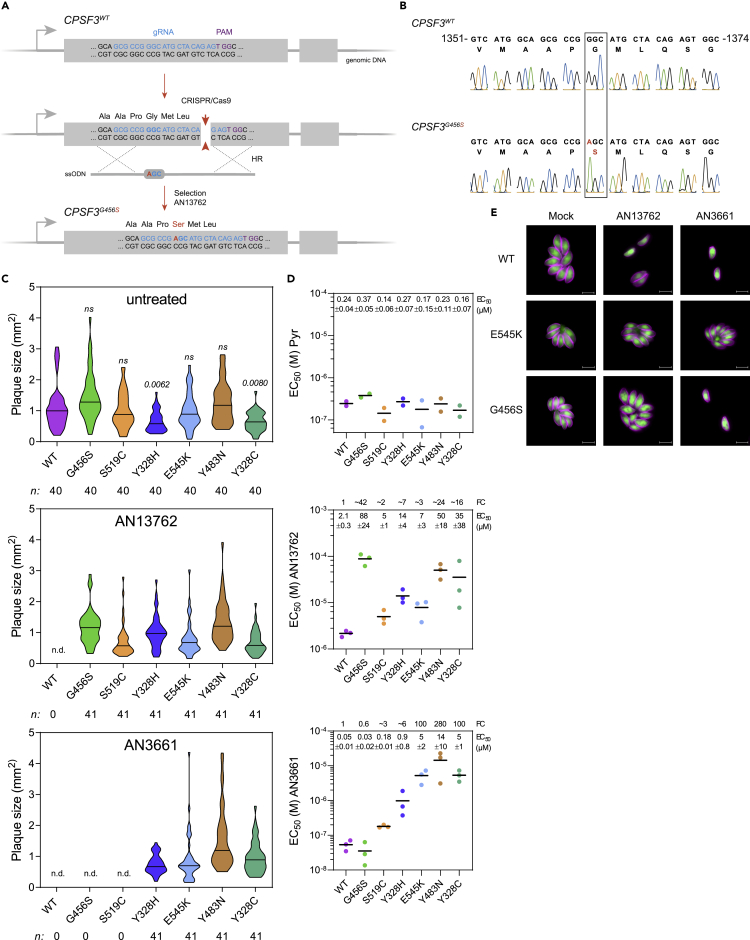
Figure 3.
Validation of T. gondii CPSF3 as the Benzoxaborole Target
(A) Schematic of the CPSF3 gene editing strategy in T. gondii parasites. Detailed view of CPSF3 locus and CRISPR/Cas9-mediated homology-directed repair with single-stranded oligo DNA nucleotides (ssODNs) carrying nucleotide substitutions (orange letters). After homologous recombination (HR) events with ssODNs, CPSF3 recombinants were selected with AN13762.
(B) Sanger sequencing validation of CPSF3 editing. Chromatograms of CPSF3 DNA sequences from parental and engineered parasites are shown. Nucleotide positions relative to the ATG start codon on genomic DNA are indicated. A complete dataset can be found in Figure S3A.
(C) Effects of CPSF3 mutations on T. gondii lytic cycle as determined by plaque assay. Plaque sizes were measured for WT and the engineered CPSF3 mutant strains (G456S, S519C, Y328H, E545K, Y483N, Y328C) after 7 days of growth in the absence or presence of 10 μM AN13762 or 5 μM AN3661. n.d., not detected. p values corresponding to Kruskal-Wallis test with Dunn's multiple comparisons with the wild-type (WT) strain are indicated. ns, not significant. Associated data are shown in Figure S3B.
(D) EC50 values for pyrimethamine (Pyr), AN13762, and AN3661 were determined for WT and the engineered CPSF3 mutant strains (G456S, S519C, Y328H, E545K, Y483N, Y328C). Data are mean from at least 2 independent biological replicates, each with 3 technical replicates. Associated dose-response curves are shown in Figure S3C. Mean EC50 values ± SD with fold changes (FC) in EC50 relative to that of the WT parasites are indicated.
(E) Fluorescence microscopy showing intracellular growth of WT and the CPSF3-edited parasites (G456S and E545K). HFF cells were infected with tachyzoites of the indicated T. gondii strains expressing the NLuc-P2A-EmGFP reporter gene and incubated with 10 μM AN13762, 5 μM AN3661, or 0.1% DMSO as control. Cells were fixed 24 h post-infection and then stained with antibodies against the T. gondii inner membrane complex protein GAP45 (magenta). The cytosolic GFP is shown in green. Scale bars, 10 μm. Complete dataset in shown in Figure S3D.