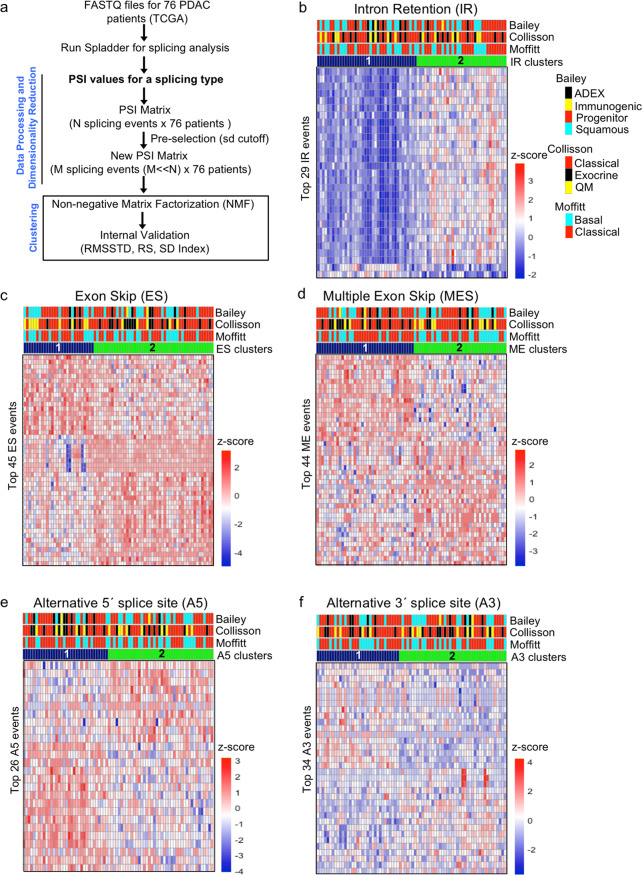
Fig. 1. Clustering of PDAC patients based on AS events.
a Workflow for NMF clustering of PDAC patients. PSI values for five types of AS events (intron retention, exon skip, multiple exon skip, and alternative 5′ and 3′ events) were obtained for 76 high-purity patients using Spladder34. The most variable events for each AS type were used for NMF clustering. The resulting clusters were compared for compactness and separation. b−f Heatmaps comparing AS levels between two patient clusters based on intron retention (b), exon skip (c), multiple exon skip (d), alternative 5′ splice site (e), and alternative 3′ splice site (f). Two clusters for each AS type were generated using the workflow shown in (a). Each column represents one patient and each row represents one of the top NMF events for that AS type. Top NMF events were obtained using the criteria described by Kim et al.96. PSI values were transformed into z-scores that are color-coded such that higher inclusion levels are shown in red and lower inclusion levels are shown in blue. Upper bars above the heat indicate the assignment of each patient to gene expression-based PDAC subtypes published by Collisson et al.12, Moffitt et al.13, and Bailey et al.14 The lowermost bar above the heatmaps represents assignment of patient clusters to a particular AS type (shown in blue and green).